| Full name: tripartite motif containing 35 | Alias Symbol: KIAA1098|MAIR|HLS5 | ||
| Type: protein-coding gene | Cytoband: 8p21.2 | ||
| Entrez ID: 23087 | HGNC ID: HGNC:16285 | Ensembl Gene: ENSG00000104228 | OMIM ID: 617007 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of TRIM35:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TRIM35 | 23087 | 227102_at | 0.3632 | 0.4277 | |
| GSE26886 | TRIM35 | 23087 | 227102_at | 0.5243 | 0.0143 | |
| GSE45670 | TRIM35 | 23087 | 227102_at | 0.1335 | 0.4073 | |
| GSE53622 | TRIM35 | 23087 | 49972 | -0.0905 | 0.3327 | |
| GSE53624 | TRIM35 | 23087 | 49972 | 0.1266 | 0.1035 | |
| GSE63941 | TRIM35 | 23087 | 227102_at | -1.3702 | 0.0036 | |
| GSE77861 | TRIM35 | 23087 | 227102_at | 0.0964 | 0.3599 | |
| GSE97050 | TRIM35 | 23087 | A_23_P502553 | 0.0426 | 0.8728 | |
| SRP007169 | TRIM35 | 23087 | RNAseq | -0.4742 | 0.2545 | |
| SRP008496 | TRIM35 | 23087 | RNAseq | -0.2677 | 0.3011 | |
| SRP064894 | TRIM35 | 23087 | RNAseq | -0.1073 | 0.4847 | |
| SRP133303 | TRIM35 | 23087 | RNAseq | -0.3061 | 0.1243 | |
| SRP159526 | TRIM35 | 23087 | RNAseq | -0.2235 | 0.4059 | |
| SRP193095 | TRIM35 | 23087 | RNAseq | -0.0189 | 0.8759 | |
| SRP219564 | TRIM35 | 23087 | RNAseq | 0.2225 | 0.5664 | |
| TCGA | TRIM35 | 23087 | RNAseq | 0.1244 | 0.0933 |
Upregulated datasets: 0; Downregulated datasets: 1.
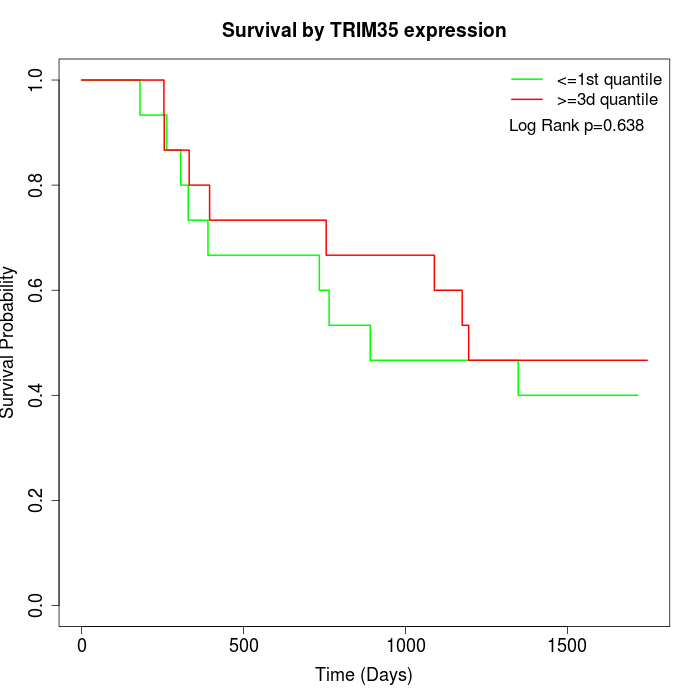
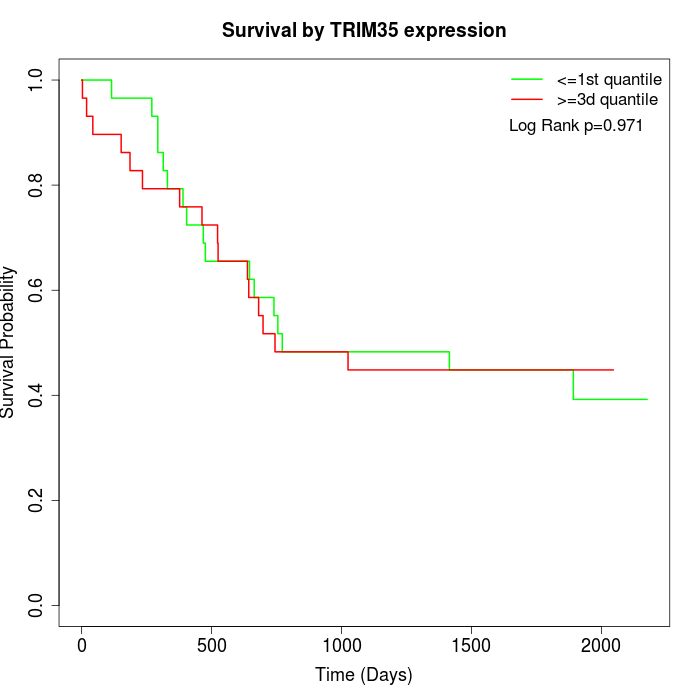
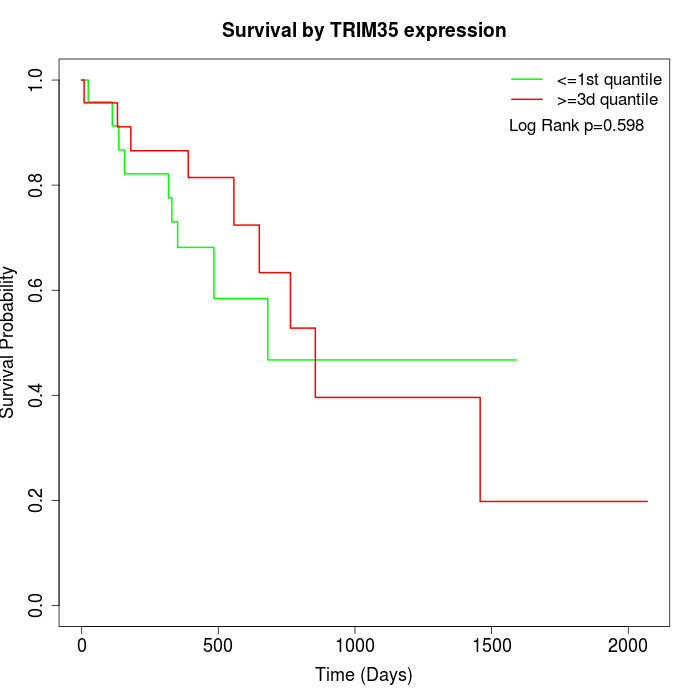
Survival by TRIM35 expression:
Note: Click image to view full size file.
Copy number change of TRIM35:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRIM35 | 23087 | 4 | 11 | 15 | |
| GSE20123 | TRIM35 | 23087 | 4 | 11 | 15 | |
| GSE43470 | TRIM35 | 23087 | 4 | 7 | 32 | |
| GSE46452 | TRIM35 | 23087 | 13 | 13 | 33 | |
| GSE47630 | TRIM35 | 23087 | 10 | 8 | 22 | |
| GSE54993 | TRIM35 | 23087 | 2 | 15 | 53 | |
| GSE54994 | TRIM35 | 23087 | 10 | 15 | 28 | |
| GSE60625 | TRIM35 | 23087 | 3 | 0 | 8 | |
| GSE74703 | TRIM35 | 23087 | 4 | 6 | 26 | |
| GSE74704 | TRIM35 | 23087 | 3 | 8 | 9 | |
| TCGA | TRIM35 | 23087 | 16 | 43 | 37 |
Total number of gains: 73; Total number of losses: 137; Total Number of normals: 278.
Somatic mutations of TRIM35:
Generating mutation plots.
Highly correlated genes for TRIM35:
Showing top 20/261 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRIM35 | CEP97 | 0.831292 | 3 | 0 | 3 |
| TRIM35 | ABHD2 | 0.805152 | 3 | 0 | 3 |
| TRIM35 | RRM2B | 0.805081 | 3 | 0 | 3 |
| TRIM35 | RRAS2 | 0.796517 | 3 | 0 | 3 |
| TRIM35 | TNPO1 | 0.790932 | 3 | 0 | 3 |
| TRIM35 | PAK2 | 0.782156 | 3 | 0 | 3 |
| TRIM35 | PUM1 | 0.776877 | 3 | 0 | 3 |
| TRIM35 | FIBP | 0.774433 | 3 | 0 | 3 |
| TRIM35 | DHX15 | 0.763942 | 3 | 0 | 3 |
| TRIM35 | USF1 | 0.760011 | 3 | 0 | 3 |
| TRIM35 | GDAP1 | 0.758718 | 3 | 0 | 3 |
| TRIM35 | OAF | 0.752599 | 3 | 0 | 3 |
| TRIM35 | SCRN1 | 0.748524 | 3 | 0 | 3 |
| TRIM35 | FAM13A | 0.74799 | 3 | 0 | 3 |
| TRIM35 | LSM10 | 0.747819 | 3 | 0 | 3 |
| TRIM35 | GAA | 0.742771 | 3 | 0 | 3 |
| TRIM35 | ACVR1 | 0.739569 | 4 | 0 | 4 |
| TRIM35 | MNAT1 | 0.731183 | 3 | 0 | 3 |
| TRIM35 | VASP | 0.728777 | 3 | 0 | 3 |
| TRIM35 | ILKAP | 0.728392 | 4 | 0 | 4 |
For details and further investigation, click here