| Full name: peptidylglycine alpha-amidating monooxygenase | Alias Symbol: PAL|PHM | ||
| Type: protein-coding gene | Cytoband: 5q21.1 | ||
| Entrez ID: 5066 | HGNC ID: HGNC:8596 | Ensembl Gene: ENSG00000145730 | OMIM ID: 170270 |
| Drug and gene relationship at DGIdb | |||
Expression of PAM:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PAM | 5066 | 202336_s_at | -0.2258 | 0.7625 | |
| GSE20347 | PAM | 5066 | 202336_s_at | 0.1325 | 0.6874 | |
| GSE23400 | PAM | 5066 | 202336_s_at | 0.1936 | 0.1559 | |
| GSE26886 | PAM | 5066 | 202336_s_at | 0.9774 | 0.0004 | |
| GSE29001 | PAM | 5066 | 202336_s_at | 0.4826 | 0.1257 | |
| GSE38129 | PAM | 5066 | 202336_s_at | -0.1260 | 0.6760 | |
| GSE45670 | PAM | 5066 | 202336_s_at | -0.5081 | 0.0461 | |
| GSE53622 | PAM | 5066 | 149457 | -0.0088 | 0.9540 | |
| GSE53624 | PAM | 5066 | 17198 | -0.0916 | 0.3665 | |
| GSE63941 | PAM | 5066 | 202336_s_at | -2.1792 | 0.0419 | |
| GSE77861 | PAM | 5066 | 202336_s_at | 0.5468 | 0.1988 | |
| GSE97050 | PAM | 5066 | A_23_P213678 | -0.3133 | 0.4765 | |
| SRP007169 | PAM | 5066 | RNAseq | -0.4483 | 0.2834 | |
| SRP008496 | PAM | 5066 | RNAseq | 0.1885 | 0.5075 | |
| SRP064894 | PAM | 5066 | RNAseq | -0.0807 | 0.7441 | |
| SRP133303 | PAM | 5066 | RNAseq | 0.3758 | 0.2160 | |
| SRP159526 | PAM | 5066 | RNAseq | 0.2605 | 0.3697 | |
| SRP193095 | PAM | 5066 | RNAseq | 0.4978 | 0.0237 | |
| SRP219564 | PAM | 5066 | RNAseq | 0.0640 | 0.9191 | |
| TCGA | PAM | 5066 | RNAseq | -0.2122 | 0.0008 |
Upregulated datasets: 0; Downregulated datasets: 1.
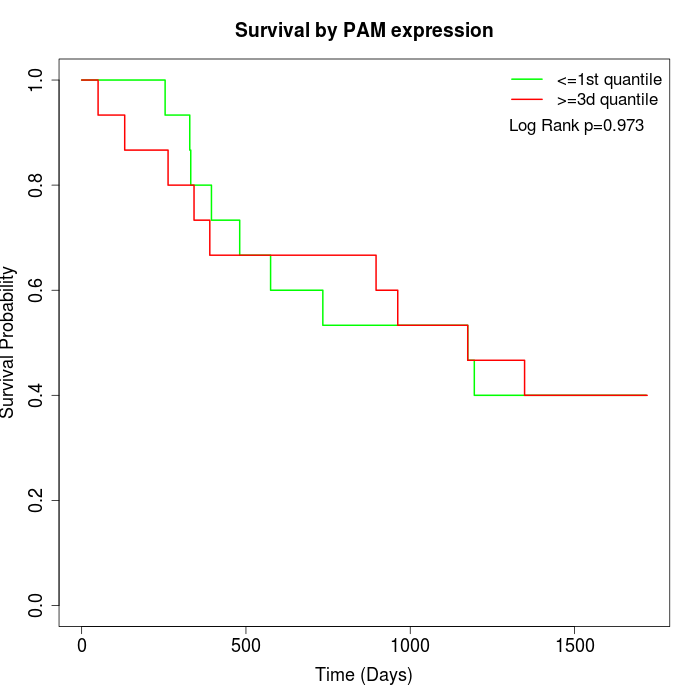
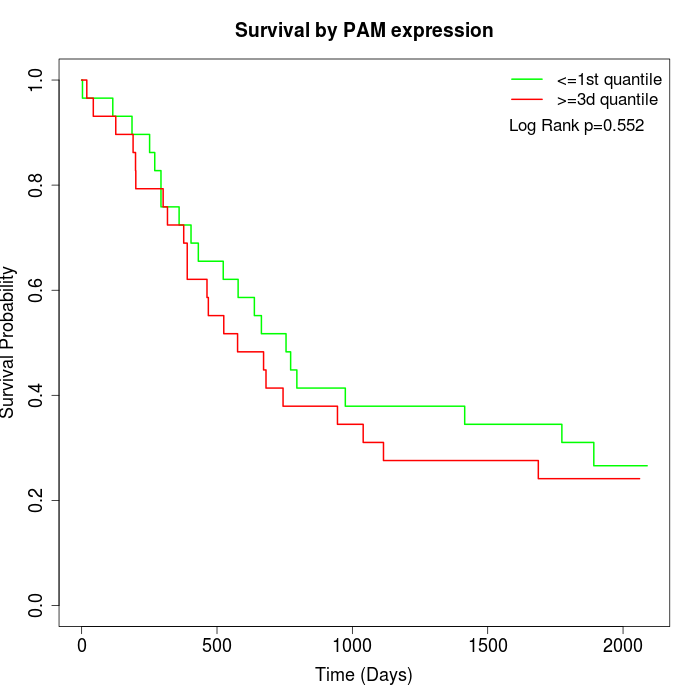
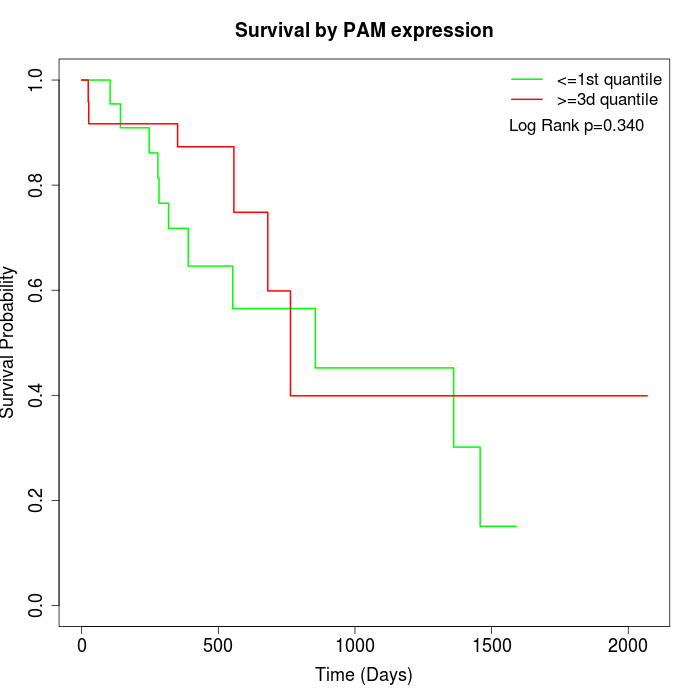
Survival by PAM expression:
Note: Click image to view full size file.
Copy number change of PAM:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PAM | 5066 | 1 | 13 | 16 | |
| GSE20123 | PAM | 5066 | 1 | 13 | 16 | |
| GSE43470 | PAM | 5066 | 3 | 9 | 31 | |
| GSE46452 | PAM | 5066 | 0 | 28 | 31 | |
| GSE47630 | PAM | 5066 | 0 | 20 | 20 | |
| GSE54993 | PAM | 5066 | 8 | 1 | 61 | |
| GSE54994 | PAM | 5066 | 1 | 17 | 35 | |
| GSE60625 | PAM | 5066 | 0 | 0 | 11 | |
| GSE74703 | PAM | 5066 | 2 | 7 | 27 | |
| GSE74704 | PAM | 5066 | 1 | 7 | 12 | |
| TCGA | PAM | 5066 | 2 | 44 | 50 |
Total number of gains: 19; Total number of losses: 159; Total Number of normals: 310.
Somatic mutations of PAM:
Generating mutation plots.
Highly correlated genes for PAM:
Showing top 20/702 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PAM | DNAJC18 | 0.755657 | 3 | 0 | 3 |
| PAM | PPP1R9A | 0.722823 | 3 | 0 | 3 |
| PAM | COL28A1 | 0.702155 | 3 | 0 | 3 |
| PAM | GNB4 | 0.696178 | 3 | 0 | 3 |
| PAM | ZC3H12C | 0.691649 | 3 | 0 | 3 |
| PAM | NKD1 | 0.67279 | 3 | 0 | 3 |
| PAM | PURA | 0.668758 | 4 | 0 | 4 |
| PAM | PDGFC | 0.663899 | 12 | 0 | 11 |
| PAM | SALL2 | 0.661779 | 3 | 0 | 3 |
| PAM | CYTH3 | 0.660432 | 6 | 0 | 6 |
| PAM | CLMP | 0.652744 | 6 | 0 | 5 |
| PAM | GPC4 | 0.650997 | 7 | 0 | 6 |
| PAM | LTBP1 | 0.65071 | 11 | 0 | 10 |
| PAM | ANO6 | 0.649296 | 8 | 0 | 7 |
| PAM | ATXN1L | 0.649218 | 3 | 0 | 3 |
| PAM | ATP11C | 0.649199 | 3 | 0 | 3 |
| PAM | RELL1 | 0.647564 | 3 | 0 | 3 |
| PAM | STX2 | 0.646468 | 11 | 0 | 9 |
| PAM | PRELID2 | 0.64526 | 3 | 0 | 3 |
| PAM | TRIB1 | 0.639992 | 4 | 0 | 4 |
For details and further investigation, click here