| Full name: G protein subunit beta 4 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 3q26.33 | ||
| Entrez ID: 59345 | HGNC ID: HGNC:20731 | Ensembl Gene: ENSG00000114450 | OMIM ID: 610863 |
| Drug and gene relationship at DGIdb | |||
GNB4 involved pathways:
Expression of GNB4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GNB4 | 59345 | 225710_at | -0.0700 | 0.9142 | |
| GSE26886 | GNB4 | 59345 | 225710_at | -0.2185 | 0.5201 | |
| GSE45670 | GNB4 | 59345 | 225710_at | -0.5025 | 0.1160 | |
| GSE63941 | GNB4 | 59345 | 225710_at | -1.8313 | 0.1673 | |
| GSE77861 | GNB4 | 59345 | 225710_at | -0.4357 | 0.5052 | |
| GSE97050 | GNB4 | 59345 | A_32_P171313 | -0.5165 | 0.1204 | |
| SRP007169 | GNB4 | 59345 | RNAseq | 2.4255 | 0.0050 | |
| SRP008496 | GNB4 | 59345 | RNAseq | 1.8113 | 0.0049 | |
| SRP064894 | GNB4 | 59345 | RNAseq | 0.6697 | 0.0043 | |
| SRP133303 | GNB4 | 59345 | RNAseq | 0.8828 | 0.0001 | |
| SRP159526 | GNB4 | 59345 | RNAseq | 0.3611 | 0.1054 | |
| SRP193095 | GNB4 | 59345 | RNAseq | 0.4558 | 0.0134 | |
| SRP219564 | GNB4 | 59345 | RNAseq | 0.1933 | 0.7364 | |
| TCGA | GNB4 | 59345 | RNAseq | 0.1399 | 0.0836 |
Upregulated datasets: 2; Downregulated datasets: 0.
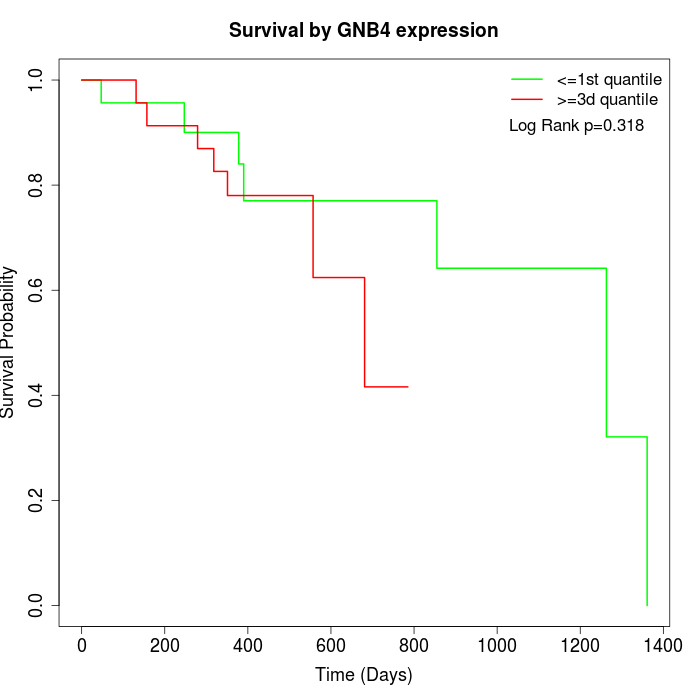
Survival by GNB4 expression:
Note: Click image to view full size file.
Copy number change of GNB4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GNB4 | 59345 | 25 | 0 | 5 | |
| GSE20123 | GNB4 | 59345 | 25 | 0 | 5 | |
| GSE43470 | GNB4 | 59345 | 24 | 0 | 19 | |
| GSE46452 | GNB4 | 59345 | 19 | 1 | 39 | |
| GSE47630 | GNB4 | 59345 | 27 | 2 | 11 | |
| GSE54993 | GNB4 | 59345 | 1 | 18 | 51 | |
| GSE54994 | GNB4 | 59345 | 41 | 0 | 12 | |
| GSE60625 | GNB4 | 59345 | 0 | 6 | 5 | |
| GSE74703 | GNB4 | 59345 | 20 | 0 | 16 | |
| GSE74704 | GNB4 | 59345 | 16 | 0 | 4 | |
| TCGA | GNB4 | 59345 | 78 | 0 | 18 |
Total number of gains: 276; Total number of losses: 27; Total Number of normals: 185.
Somatic mutations of GNB4:
Generating mutation plots.
Highly correlated genes for GNB4:
Showing top 20/227 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GNB4 | ERRFI1 | 0.794035 | 3 | 0 | 3 |
| GNB4 | SNX14 | 0.783637 | 3 | 0 | 3 |
| GNB4 | GPM6B | 0.780535 | 3 | 0 | 3 |
| GNB4 | FAM122C | 0.767682 | 3 | 0 | 3 |
| GNB4 | SNTB1 | 0.765315 | 3 | 0 | 3 |
| GNB4 | TOB2 | 0.748602 | 3 | 0 | 3 |
| GNB4 | CLMP | 0.747803 | 4 | 0 | 4 |
| GNB4 | C5orf24 | 0.744257 | 3 | 0 | 3 |
| GNB4 | SRPX | 0.742332 | 3 | 0 | 3 |
| GNB4 | KIF5B | 0.741912 | 3 | 0 | 3 |
| GNB4 | ILK | 0.736536 | 3 | 0 | 3 |
| GNB4 | PPP1R15B | 0.734933 | 3 | 0 | 3 |
| GNB4 | STARD3NL | 0.73371 | 3 | 0 | 3 |
| GNB4 | ME3 | 0.727444 | 3 | 0 | 3 |
| GNB4 | STX2 | 0.72395 | 4 | 0 | 4 |
| GNB4 | WBP4 | 0.722221 | 3 | 0 | 3 |
| GNB4 | OPTN | 0.718884 | 3 | 0 | 3 |
| GNB4 | DIS3L2 | 0.718785 | 3 | 0 | 3 |
| GNB4 | LSM11 | 0.717194 | 4 | 0 | 3 |
| GNB4 | SRSF7 | 0.714146 | 3 | 0 | 3 |
For details and further investigation, click here