| Full name: phosphodiesterase 6G | Alias Symbol: RP57 | ||
| Type: protein-coding gene | Cytoband: 17q25.3 | ||
| Entrez ID: 5148 | HGNC ID: HGNC:8789 | Ensembl Gene: ENSG00000185527 | OMIM ID: 180073 |
| Drug and gene relationship at DGIdb | |||
Expression of PDE6G:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PDE6G | 5148 | 210060_at | -0.1559 | 0.5706 | |
| GSE20347 | PDE6G | 5148 | 210060_at | -0.0271 | 0.8107 | |
| GSE23400 | PDE6G | 5148 | 210060_at | -0.1090 | 0.0012 | |
| GSE26886 | PDE6G | 5148 | 210060_at | -0.0490 | 0.7713 | |
| GSE29001 | PDE6G | 5148 | 210060_at | -0.1738 | 0.3222 | |
| GSE38129 | PDE6G | 5148 | 210060_at | -0.1584 | 0.0487 | |
| GSE45670 | PDE6G | 5148 | 210060_at | 0.1538 | 0.1100 | |
| GSE63941 | PDE6G | 5148 | 210060_at | 0.2539 | 0.0687 | |
| GSE77861 | PDE6G | 5148 | 210060_at | -0.1850 | 0.0754 | |
| TCGA | PDE6G | 5148 | RNAseq | 0.2442 | 0.5392 |
Upregulated datasets: 0; Downregulated datasets: 0.
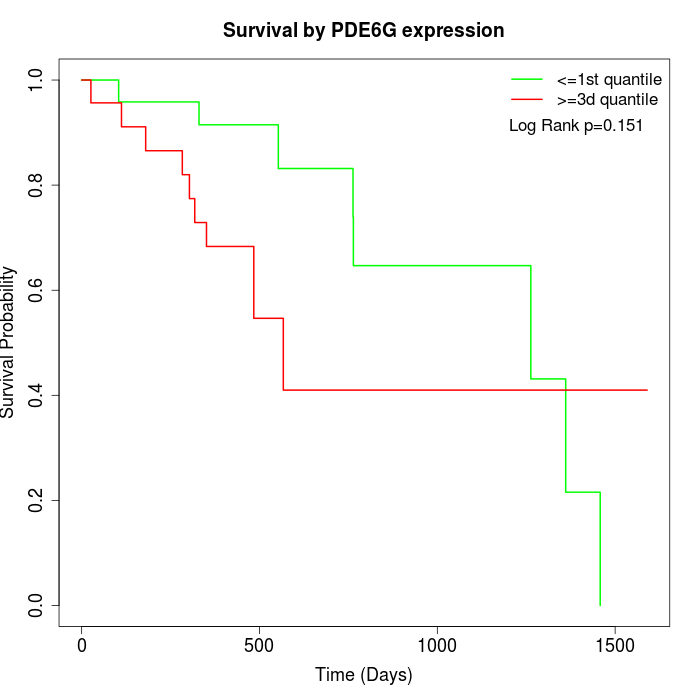
Survival by PDE6G expression:
Note: Click image to view full size file.
Copy number change of PDE6G:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PDE6G | 5148 | 4 | 4 | 22 | |
| GSE20123 | PDE6G | 5148 | 4 | 4 | 22 | |
| GSE43470 | PDE6G | 5148 | 3 | 5 | 35 | |
| GSE46452 | PDE6G | 5148 | 34 | 0 | 25 | |
| GSE47630 | PDE6G | 5148 | 9 | 2 | 29 | |
| GSE54993 | PDE6G | 5148 | 2 | 5 | 63 | |
| GSE54994 | PDE6G | 5148 | 10 | 6 | 37 | |
| GSE60625 | PDE6G | 5148 | 6 | 0 | 5 | |
| GSE74703 | PDE6G | 5148 | 3 | 3 | 30 | |
| GSE74704 | PDE6G | 5148 | 4 | 3 | 13 | |
| TCGA | PDE6G | 5148 | 25 | 12 | 59 |
Total number of gains: 104; Total number of losses: 44; Total Number of normals: 340.
Somatic mutations of PDE6G:
Generating mutation plots.
Highly correlated genes for PDE6G:
Showing top 20/1042 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PDE6G | ANKRD31 | 0.732044 | 3 | 0 | 3 |
| PDE6G | SNX11 | 0.715642 | 3 | 0 | 3 |
| PDE6G | CYP4A11 | 0.707802 | 7 | 0 | 7 |
| PDE6G | LINC01210 | 0.68205 | 4 | 0 | 3 |
| PDE6G | OR4D2 | 0.679264 | 4 | 0 | 3 |
| PDE6G | LINC00930 | 0.677737 | 4 | 0 | 4 |
| PDE6G | CEACAM21 | 0.673939 | 8 | 0 | 7 |
| PDE6G | LINC00112 | 0.672725 | 3 | 0 | 3 |
| PDE6G | EPOR | 0.665972 | 7 | 0 | 6 |
| PDE6G | OR51M1 | 0.654948 | 5 | 0 | 5 |
| PDE6G | CBLN4 | 0.654112 | 4 | 0 | 4 |
| PDE6G | ANHX | 0.654054 | 3 | 0 | 3 |
| PDE6G | NAAA | 0.652839 | 3 | 0 | 3 |
| PDE6G | BPIFA1 | 0.65237 | 6 | 0 | 6 |
| PDE6G | AVPR1B | 0.646259 | 9 | 0 | 8 |
| PDE6G | STH | 0.645331 | 4 | 0 | 4 |
| PDE6G | NEUROD4 | 0.643948 | 6 | 0 | 5 |
| PDE6G | MUC5AC | 0.642412 | 5 | 0 | 5 |
| PDE6G | CA1 | 0.641932 | 7 | 0 | 7 |
| PDE6G | ZGLP1 | 0.641916 | 5 | 0 | 4 |
For details and further investigation, click here