| Full name: cerebellin 4 precursor | Alias Symbol: dJ885A10.1 | ||
| Type: protein-coding gene | Cytoband: 20q13.2 | ||
| Entrez ID: 140689 | HGNC ID: HGNC:16231 | Ensembl Gene: ENSG00000054803 | OMIM ID: 615029 |
| Drug and gene relationship at DGIdb | |||
Expression of CBLN4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CBLN4 | 140689 | 234024_at | -0.0625 | 0.8706 | |
| GSE26886 | CBLN4 | 140689 | 234024_at | -0.0302 | 0.7960 | |
| GSE45670 | CBLN4 | 140689 | 234024_at | 0.0037 | 0.9800 | |
| GSE53622 | CBLN4 | 140689 | 86678 | 0.2042 | 0.0357 | |
| GSE53624 | CBLN4 | 140689 | 86678 | 0.2871 | 0.0007 | |
| GSE63941 | CBLN4 | 140689 | 234024_at | 0.0158 | 0.9306 | |
| GSE77861 | CBLN4 | 140689 | 234024_at | -0.3103 | 0.0072 | |
| GSE97050 | CBLN4 | 140689 | A_33_P3385516 | 0.0945 | 0.6782 | |
| TCGA | CBLN4 | 140689 | RNAseq | 0.0373 | 0.9614 |
Upregulated datasets: 0; Downregulated datasets: 0.
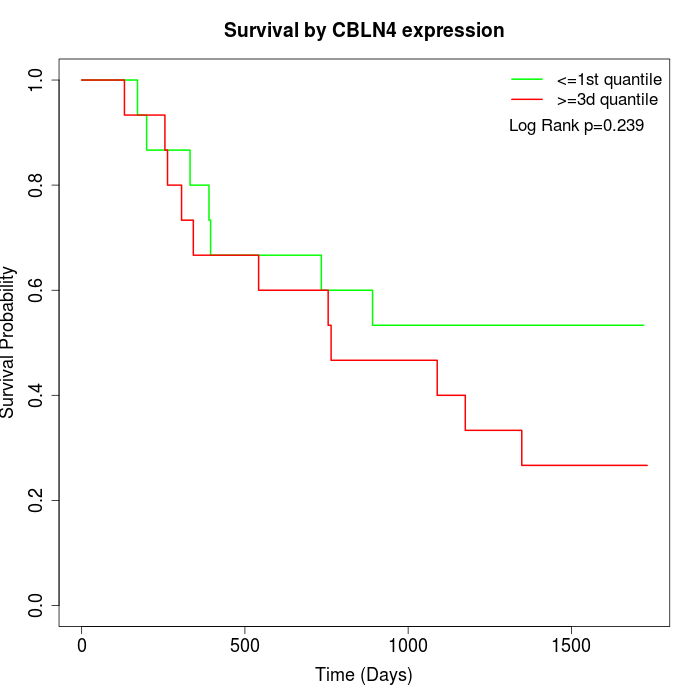
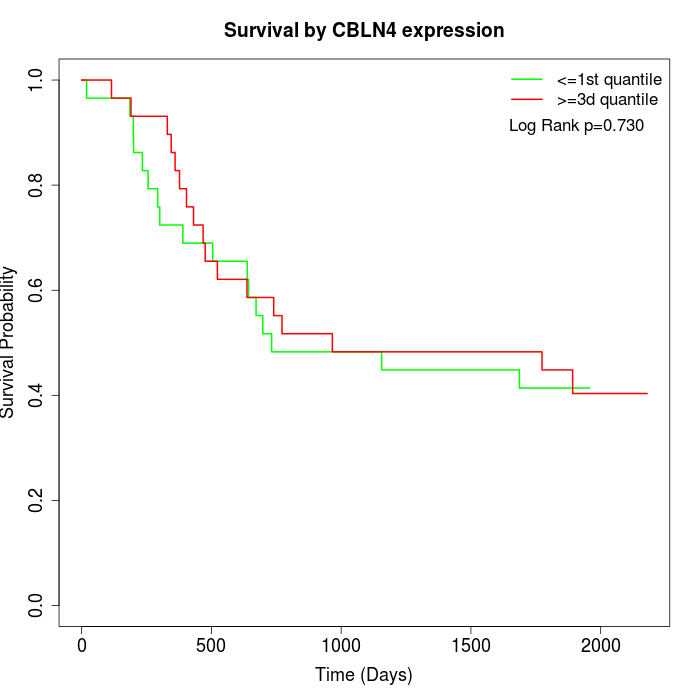
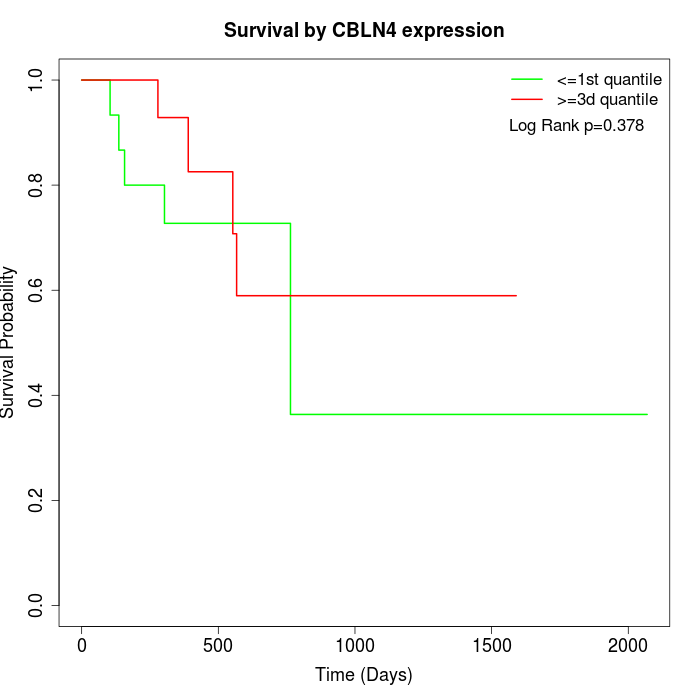
Survival by CBLN4 expression:
Note: Click image to view full size file.
Copy number change of CBLN4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CBLN4 | 140689 | 15 | 1 | 14 | |
| GSE20123 | CBLN4 | 140689 | 15 | 1 | 14 | |
| GSE43470 | CBLN4 | 140689 | 13 | 0 | 30 | |
| GSE46452 | CBLN4 | 140689 | 29 | 0 | 30 | |
| GSE47630 | CBLN4 | 140689 | 24 | 1 | 15 | |
| GSE54993 | CBLN4 | 140689 | 0 | 17 | 53 | |
| GSE54994 | CBLN4 | 140689 | 27 | 0 | 26 | |
| GSE60625 | CBLN4 | 140689 | 0 | 0 | 11 | |
| GSE74703 | CBLN4 | 140689 | 11 | 0 | 25 | |
| GSE74704 | CBLN4 | 140689 | 12 | 0 | 8 | |
| TCGA | CBLN4 | 140689 | 44 | 4 | 48 |
Total number of gains: 190; Total number of losses: 24; Total Number of normals: 274.
Somatic mutations of CBLN4:
Generating mutation plots.
Highly correlated genes for CBLN4:
Showing top 20/404 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CBLN4 | CCDC42 | 0.781536 | 3 | 0 | 3 |
| CBLN4 | RIPK4 | 0.767011 | 3 | 0 | 3 |
| CBLN4 | GUCY2C | 0.766426 | 3 | 0 | 3 |
| CBLN4 | VGLL2 | 0.733607 | 3 | 0 | 3 |
| CBLN4 | KRT35 | 0.726062 | 3 | 0 | 3 |
| CBLN4 | RETN | 0.721962 | 3 | 0 | 3 |
| CBLN4 | MTUS2-AS1 | 0.713278 | 3 | 0 | 3 |
| CBLN4 | MAST4 | 0.707858 | 3 | 0 | 3 |
| CBLN4 | PTK6 | 0.707705 | 3 | 0 | 3 |
| CBLN4 | PKLR | 0.706903 | 3 | 0 | 3 |
| CBLN4 | SHISA7 | 0.704187 | 3 | 0 | 3 |
| CBLN4 | CCL25 | 0.7028 | 4 | 0 | 4 |
| CBLN4 | EPS8L2 | 0.700991 | 3 | 0 | 3 |
| CBLN4 | CACNA1G | 0.699652 | 4 | 0 | 4 |
| CBLN4 | TERT | 0.694156 | 4 | 0 | 3 |
| CBLN4 | FAM189A2 | 0.691827 | 3 | 0 | 3 |
| CBLN4 | YIPF1 | 0.688793 | 3 | 0 | 3 |
| CBLN4 | BBOX1 | 0.688766 | 3 | 0 | 3 |
| CBLN4 | HCG9 | 0.688704 | 3 | 0 | 3 |
| CBLN4 | GALR3 | 0.687633 | 4 | 0 | 3 |
For details and further investigation, click here