| Full name: pyruvate dehydrogenase E1 alpha 2 subunit | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 4q22.3 | ||
| Entrez ID: 5161 | HGNC ID: HGNC:8807 | Ensembl Gene: ENSG00000163114 | OMIM ID: 179061 |
| Drug and gene relationship at DGIdb | |||
PDHA2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04066 | HIF-1 signaling pathway |
Expression of PDHA2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PDHA2 | 5161 | 214518_at | -0.0713 | 0.7952 | |
| GSE20347 | PDHA2 | 5161 | 214518_at | -0.0471 | 0.5041 | |
| GSE23400 | PDHA2 | 5161 | 214518_at | -0.1913 | 0.0000 | |
| GSE26886 | PDHA2 | 5161 | 214518_at | -0.0762 | 0.5184 | |
| GSE29001 | PDHA2 | 5161 | 214518_at | -0.0395 | 0.7626 | |
| GSE38129 | PDHA2 | 5161 | 214518_at | -0.1342 | 0.0193 | |
| GSE45670 | PDHA2 | 5161 | 214518_at | 0.0930 | 0.3143 | |
| GSE53622 | PDHA2 | 5161 | 67025 | 0.3224 | 0.0001 | |
| GSE53624 | PDHA2 | 5161 | 67025 | 0.1347 | 0.5639 | |
| GSE63941 | PDHA2 | 5161 | 214518_at | 0.0238 | 0.8967 | |
| GSE77861 | PDHA2 | 5161 | 214518_at | -0.0464 | 0.7240 |
Upregulated datasets: 0; Downregulated datasets: 0.
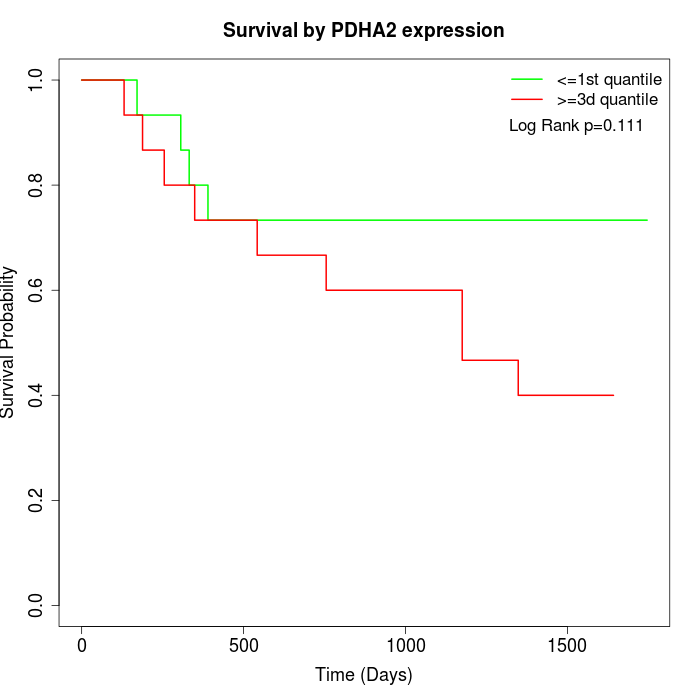
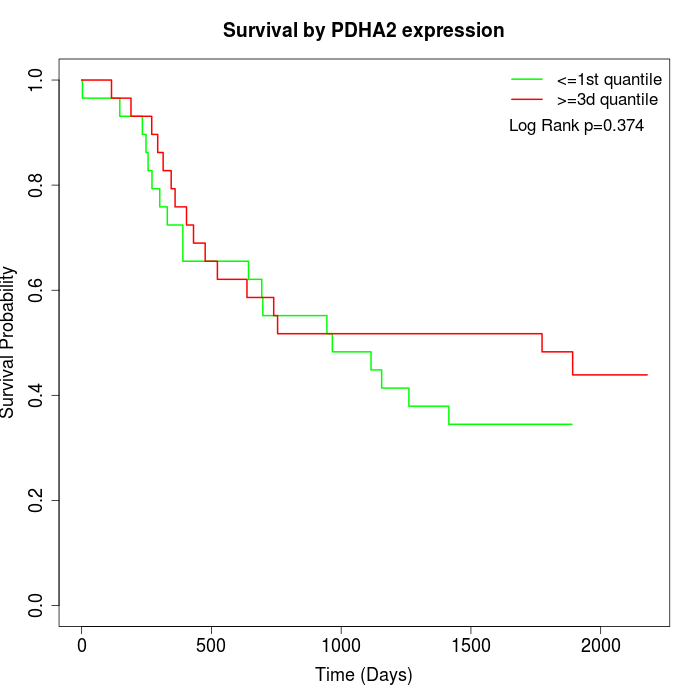
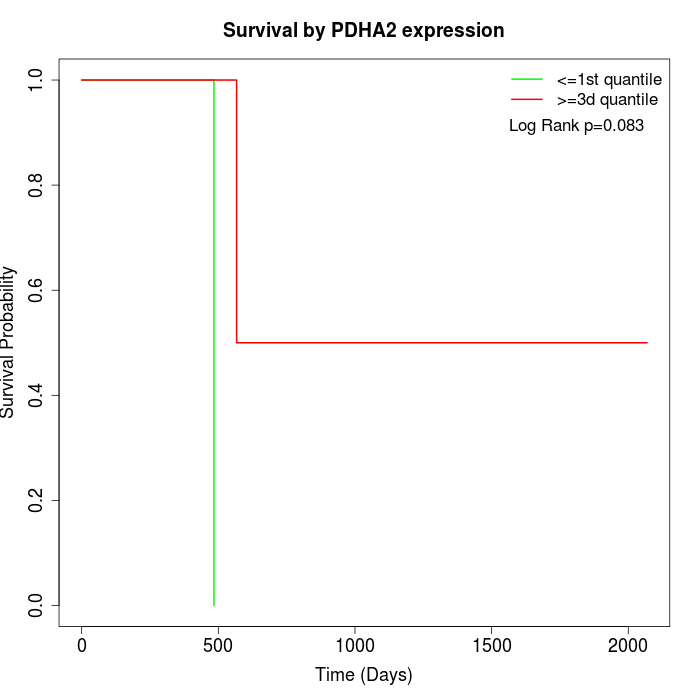
Survival by PDHA2 expression:
Note: Click image to view full size file.
Copy number change of PDHA2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PDHA2 | 5161 | 0 | 12 | 18 | |
| GSE20123 | PDHA2 | 5161 | 0 | 12 | 18 | |
| GSE43470 | PDHA2 | 5161 | 0 | 15 | 28 | |
| GSE46452 | PDHA2 | 5161 | 1 | 36 | 22 | |
| GSE47630 | PDHA2 | 5161 | 0 | 20 | 20 | |
| GSE54993 | PDHA2 | 5161 | 8 | 0 | 62 | |
| GSE54994 | PDHA2 | 5161 | 1 | 11 | 41 | |
| GSE60625 | PDHA2 | 5161 | 0 | 3 | 8 | |
| GSE74703 | PDHA2 | 5161 | 0 | 13 | 23 | |
| GSE74704 | PDHA2 | 5161 | 0 | 6 | 14 | |
| TCGA | PDHA2 | 5161 | 9 | 39 | 48 |
Total number of gains: 19; Total number of losses: 167; Total Number of normals: 302.
Somatic mutations of PDHA2:
Generating mutation plots.
Highly correlated genes for PDHA2:
Showing top 20/440 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PDHA2 | OXTR | 0.693097 | 3 | 0 | 3 |
| PDHA2 | DAOA | 0.692158 | 3 | 0 | 3 |
| PDHA2 | CIB3 | 0.68984 | 3 | 0 | 3 |
| PDHA2 | BRSK2 | 0.677177 | 3 | 0 | 3 |
| PDHA2 | TMEM151B | 0.674655 | 6 | 0 | 6 |
| PDHA2 | PARD6G-AS1 | 0.663404 | 3 | 0 | 3 |
| PDHA2 | C8orf74 | 0.656735 | 3 | 0 | 3 |
| PDHA2 | FAM222A-AS1 | 0.654601 | 3 | 0 | 3 |
| PDHA2 | PNPLA7 | 0.654109 | 3 | 0 | 3 |
| PDHA2 | ALPI | 0.641547 | 4 | 0 | 3 |
| PDHA2 | OR11A1 | 0.636513 | 5 | 0 | 4 |
| PDHA2 | FAM131C | 0.633191 | 3 | 0 | 3 |
| PDHA2 | KCNJ9 | 0.631221 | 4 | 0 | 3 |
| PDHA2 | C1orf167 | 0.629181 | 3 | 0 | 3 |
| PDHA2 | LINC01432 | 0.62037 | 3 | 0 | 3 |
| PDHA2 | RAX2 | 0.618877 | 5 | 0 | 4 |
| PDHA2 | GJD2 | 0.618762 | 4 | 0 | 3 |
| PDHA2 | WNT16 | 0.617518 | 4 | 0 | 3 |
| PDHA2 | KCNS2 | 0.616353 | 3 | 0 | 3 |
| PDHA2 | PAX8 | 0.616296 | 4 | 0 | 3 |
For details and further investigation, click here