| Full name: potassium inwardly rectifying channel subfamily J member 9 | Alias Symbol: Kir3.3|GIRK3 | ||
| Type: protein-coding gene | Cytoband: 1q23.2 | ||
| Entrez ID: 3765 | HGNC ID: HGNC:6270 | Ensembl Gene: ENSG00000162728 | OMIM ID: 600932 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
KCNJ9 involved pathways:
Expression of KCNJ9:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNJ9 | 3765 | 207527_at | 0.0475 | 0.8532 | |
| GSE20347 | KCNJ9 | 3765 | 207527_at | 0.0794 | 0.2283 | |
| GSE23400 | KCNJ9 | 3765 | 207527_at | -0.0751 | 0.0663 | |
| GSE26886 | KCNJ9 | 3765 | 207527_at | -0.0012 | 0.9930 | |
| GSE29001 | KCNJ9 | 3765 | 207527_at | -0.0716 | 0.5803 | |
| GSE38129 | KCNJ9 | 3765 | 207527_at | -0.0136 | 0.8551 | |
| GSE45670 | KCNJ9 | 3765 | 207527_at | -0.0247 | 0.8103 | |
| GSE53622 | KCNJ9 | 3765 | 134682 | 0.4791 | 0.0037 | |
| GSE53624 | KCNJ9 | 3765 | 134682 | 0.1141 | 0.6081 | |
| GSE63941 | KCNJ9 | 3765 | 207527_at | 0.0515 | 0.7485 | |
| GSE77861 | KCNJ9 | 3765 | 207527_at | 0.0575 | 0.6869 | |
| SRP133303 | KCNJ9 | 3765 | RNAseq | -1.1310 | 0.0003 | |
| TCGA | KCNJ9 | 3765 | RNAseq | -0.3693 | 0.6168 |
Upregulated datasets: 0; Downregulated datasets: 1.
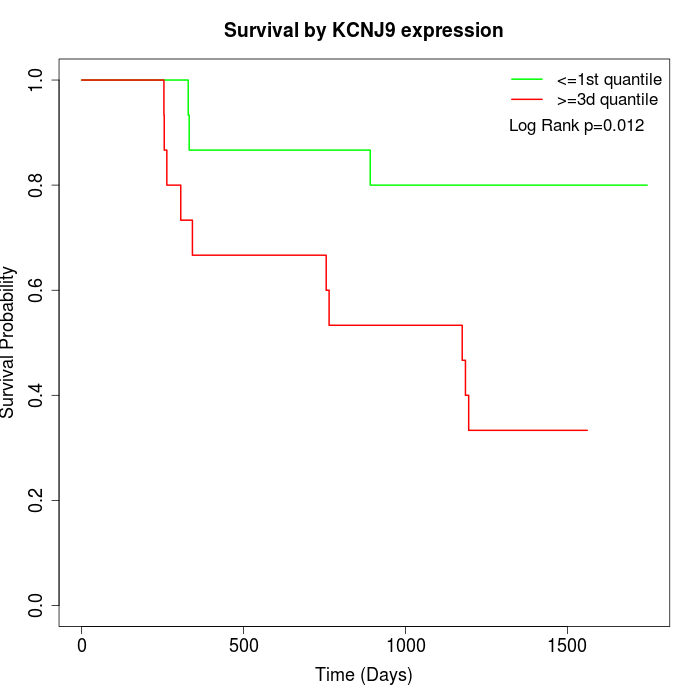
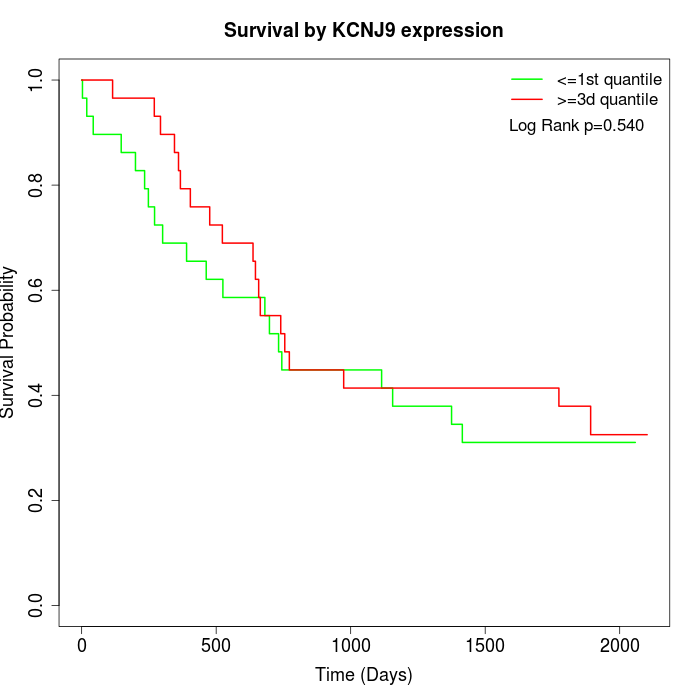
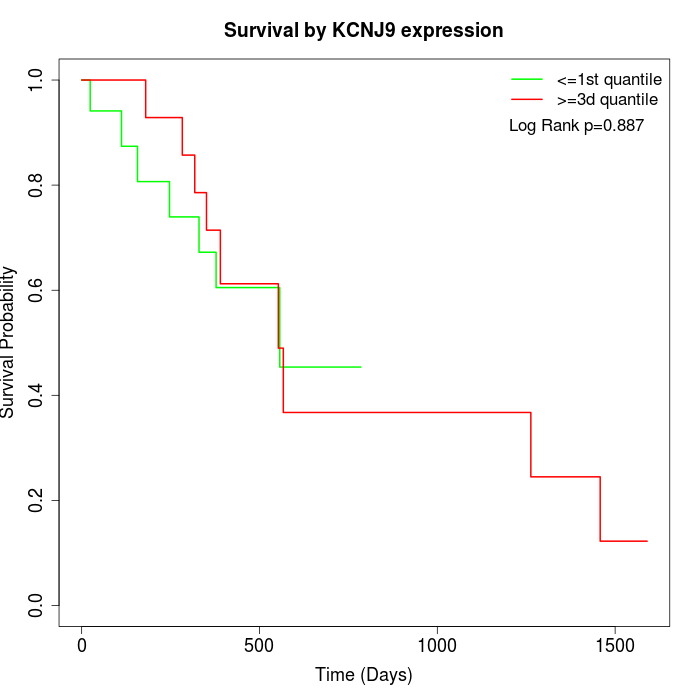
Survival by KCNJ9 expression:
Note: Click image to view full size file.
Copy number change of KCNJ9:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNJ9 | 3765 | 10 | 0 | 20 | |
| GSE20123 | KCNJ9 | 3765 | 10 | 0 | 20 | |
| GSE43470 | KCNJ9 | 3765 | 7 | 2 | 34 | |
| GSE46452 | KCNJ9 | 3765 | 2 | 1 | 56 | |
| GSE47630 | KCNJ9 | 3765 | 15 | 0 | 25 | |
| GSE54993 | KCNJ9 | 3765 | 0 | 5 | 65 | |
| GSE54994 | KCNJ9 | 3765 | 16 | 0 | 37 | |
| GSE60625 | KCNJ9 | 3765 | 0 | 0 | 11 | |
| GSE74703 | KCNJ9 | 3765 | 7 | 2 | 27 | |
| GSE74704 | KCNJ9 | 3765 | 4 | 0 | 16 | |
| TCGA | KCNJ9 | 3765 | 44 | 3 | 49 |
Total number of gains: 115; Total number of losses: 13; Total Number of normals: 360.
Somatic mutations of KCNJ9:
Generating mutation plots.
Highly correlated genes for KCNJ9:
Showing top 20/838 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNJ9 | SLC25A53 | 0.750531 | 3 | 0 | 3 |
| KCNJ9 | LRRTM1 | 0.746517 | 3 | 0 | 3 |
| KCNJ9 | VAX2 | 0.734828 | 4 | 0 | 4 |
| KCNJ9 | TTTY5 | 0.734647 | 3 | 0 | 3 |
| KCNJ9 | RPS6KA6 | 0.722782 | 3 | 0 | 3 |
| KCNJ9 | CDRT15 | 0.71848 | 4 | 0 | 4 |
| KCNJ9 | SPACA3 | 0.711492 | 3 | 0 | 3 |
| KCNJ9 | DEFB132 | 0.71145 | 3 | 0 | 3 |
| KCNJ9 | CRHR2 | 0.708103 | 6 | 0 | 5 |
| KCNJ9 | CHMP6 | 0.707328 | 3 | 0 | 3 |
| KCNJ9 | SYT9 | 0.705735 | 3 | 0 | 3 |
| KCNJ9 | TNNI1 | 0.700127 | 5 | 0 | 5 |
| KCNJ9 | CDH24 | 0.699719 | 4 | 0 | 3 |
| KCNJ9 | SUN5 | 0.699437 | 3 | 0 | 3 |
| KCNJ9 | GPSM1 | 0.696581 | 3 | 0 | 3 |
| KCNJ9 | SIRPB1 | 0.694498 | 6 | 0 | 5 |
| KCNJ9 | TMEM105 | 0.694241 | 3 | 0 | 3 |
| KCNJ9 | LINC00408 | 0.691995 | 4 | 0 | 4 |
| KCNJ9 | TMEM63B | 0.690328 | 3 | 0 | 3 |
| KCNJ9 | LGALS12 | 0.688712 | 3 | 0 | 3 |
For details and further investigation, click here