| Full name: potassium voltage-gated channel modifier subfamily S member 2 | Alias Symbol: Kv9.2 | ||
| Type: protein-coding gene | Cytoband: 8q22.2 | ||
| Entrez ID: 3788 | HGNC ID: HGNC:6301 | Ensembl Gene: ENSG00000156486 | OMIM ID: 602906 |
| Drug and gene relationship at DGIdb | |||
Expression of KCNS2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KCNS2 | 3788 | 1554212_s_at | -0.0078 | 0.9813 | |
| GSE26886 | KCNS2 | 3788 | 1554212_s_at | -0.1691 | 0.2481 | |
| GSE45670 | KCNS2 | 3788 | 1554212_s_at | -0.0884 | 0.3270 | |
| GSE53622 | KCNS2 | 3788 | 63151 | 0.8856 | 0.0000 | |
| GSE53624 | KCNS2 | 3788 | 63151 | 0.7579 | 0.0000 | |
| GSE63941 | KCNS2 | 3788 | 1554212_s_at | -0.1297 | 0.3256 | |
| GSE77861 | KCNS2 | 3788 | 1554212_s_at | -0.0802 | 0.4558 | |
| GSE97050 | KCNS2 | 3788 | A_24_P46911 | 0.6116 | 0.1443 | |
| SRP133303 | KCNS2 | 3788 | RNAseq | 0.6652 | 0.0146 | |
| TCGA | KCNS2 | 3788 | RNAseq | -2.3790 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
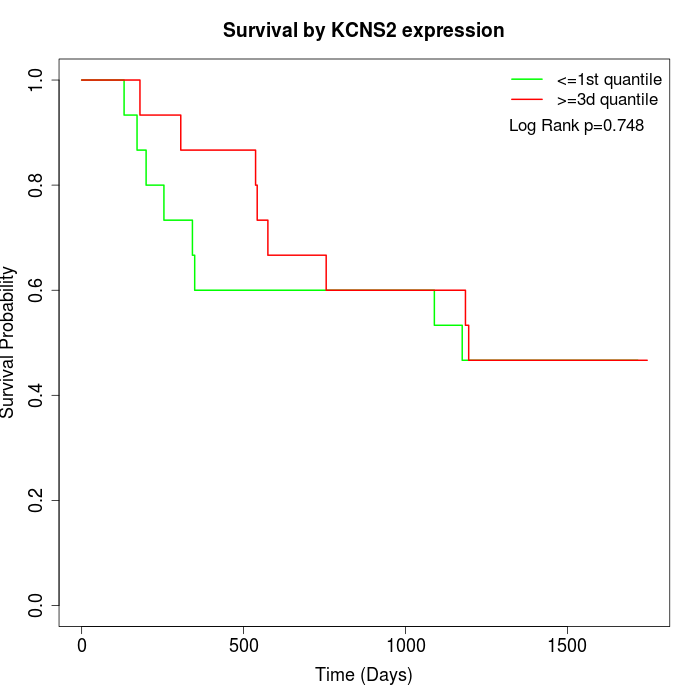
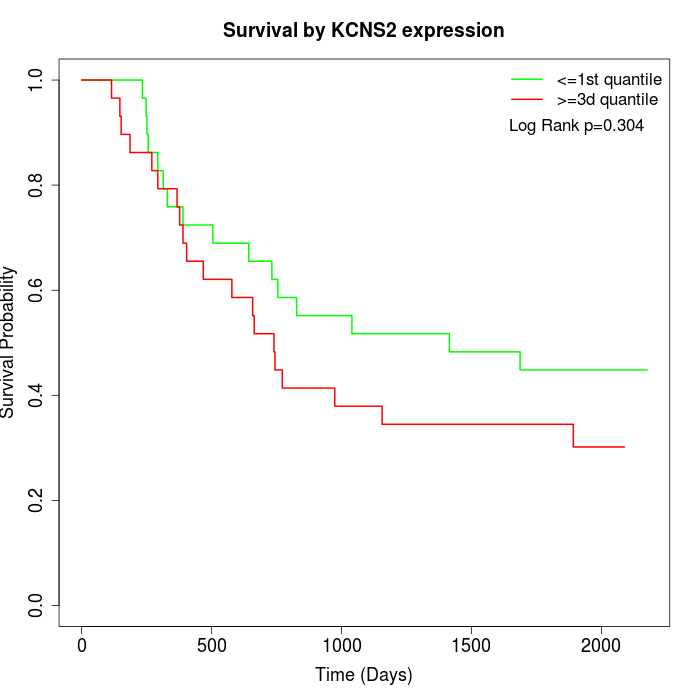
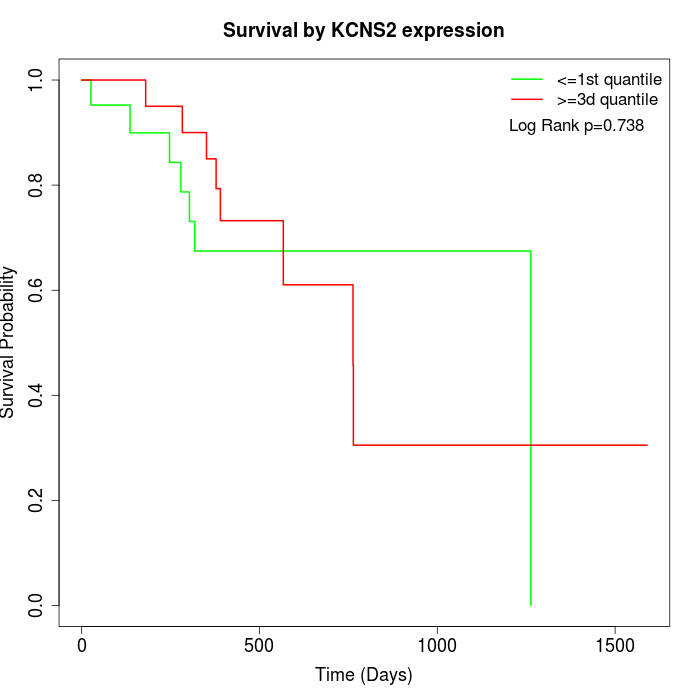
Survival by KCNS2 expression:
Note: Click image to view full size file.
Copy number change of KCNS2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCNS2 | 3788 | 16 | 0 | 14 | |
| GSE20123 | KCNS2 | 3788 | 17 | 0 | 13 | |
| GSE43470 | KCNS2 | 3788 | 21 | 1 | 21 | |
| GSE46452 | KCNS2 | 3788 | 23 | 0 | 36 | |
| GSE47630 | KCNS2 | 3788 | 24 | 0 | 16 | |
| GSE54993 | KCNS2 | 3788 | 0 | 20 | 50 | |
| GSE54994 | KCNS2 | 3788 | 37 | 1 | 15 | |
| GSE60625 | KCNS2 | 3788 | 0 | 4 | 7 | |
| GSE74703 | KCNS2 | 3788 | 18 | 1 | 17 | |
| GSE74704 | KCNS2 | 3788 | 12 | 0 | 8 | |
| TCGA | KCNS2 | 3788 | 55 | 2 | 39 |
Total number of gains: 223; Total number of losses: 29; Total Number of normals: 236.
Somatic mutations of KCNS2:
Generating mutation plots.
Highly correlated genes for KCNS2:
Showing top 20/116 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCNS2 | AUP1 | 0.730457 | 3 | 0 | 3 |
| KCNS2 | IGF2BP2 | 0.724364 | 3 | 0 | 3 |
| KCNS2 | ARFGAP1 | 0.719099 | 3 | 0 | 3 |
| KCNS2 | SLC19A1 | 0.715331 | 3 | 0 | 3 |
| KCNS2 | EXOC3L2 | 0.71282 | 3 | 0 | 3 |
| KCNS2 | SIX1 | 0.704719 | 3 | 0 | 3 |
| KCNS2 | HOXD11 | 0.700102 | 3 | 0 | 3 |
| KCNS2 | NCLN | 0.691543 | 3 | 0 | 3 |
| KCNS2 | FSCN1 | 0.690785 | 3 | 0 | 3 |
| KCNS2 | TTYH3 | 0.690724 | 3 | 0 | 3 |
| KCNS2 | ANKLE2 | 0.688876 | 3 | 0 | 3 |
| KCNS2 | PSMD2 | 0.674599 | 3 | 0 | 3 |
| KCNS2 | LMNB2 | 0.673273 | 3 | 0 | 3 |
| KCNS2 | SLC39A4 | 0.668646 | 3 | 0 | 3 |
| KCNS2 | CDC25B | 0.66799 | 3 | 0 | 3 |
| KCNS2 | DNAJB11 | 0.667317 | 3 | 0 | 3 |
| KCNS2 | CHPF | 0.665321 | 3 | 0 | 3 |
| KCNS2 | CENPI | 0.660665 | 3 | 0 | 3 |
| KCNS2 | TSPO2 | 0.656114 | 3 | 0 | 3 |
| KCNS2 | MMP1 | 0.654328 | 3 | 0 | 3 |
For details and further investigation, click here