| Full name: placental growth factor | Alias Symbol: PLGF|PlGF-2|PlGF|SHGC-10760|D12S1900|PIGF | ||
| Type: protein-coding gene | Cytoband: 14q24.3 | ||
| Entrez ID: 5228 | HGNC ID: HGNC:8893 | Ensembl Gene: ENSG00000119630 | OMIM ID: 601121 |
| Drug and gene relationship at DGIdb | |||
PGF involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04014 | Ras signaling pathway | |
| hsa04015 | Rap1 signaling pathway | |
| hsa04151 | PI3K-Akt signaling pathway | |
| hsa04510 | Focal adhesion | |
| hsa05200 | Pathways in cancer |
Expression of PGF:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PGF | 5228 | 215179_x_at | 0.0976 | 0.9044 | |
| GSE20347 | PGF | 5228 | 215179_x_at | 0.3301 | 0.1289 | |
| GSE23400 | PGF | 5228 | 215179_x_at | -0.0276 | 0.7787 | |
| GSE26886 | PGF | 5228 | 215179_x_at | 0.0810 | 0.7261 | |
| GSE29001 | PGF | 5228 | 215179_x_at | 0.1603 | 0.4001 | |
| GSE38129 | PGF | 5228 | 215179_x_at | 0.0782 | 0.7823 | |
| GSE45670 | PGF | 5228 | 215179_x_at | 0.2624 | 0.0228 | |
| GSE53622 | PGF | 5228 | 80555 | 1.7450 | 0.0000 | |
| GSE53624 | PGF | 5228 | 127285 | 1.4343 | 0.0000 | |
| GSE63941 | PGF | 5228 | 215179_x_at | -0.2491 | 0.2006 | |
| GSE77861 | PGF | 5228 | 215179_x_at | 0.6778 | 0.1344 | |
| GSE97050 | PGF | 5228 | A_33_P3406778 | 0.0332 | 0.8765 | |
| SRP007169 | PGF | 5228 | RNAseq | 3.5085 | 0.0000 | |
| SRP008496 | PGF | 5228 | RNAseq | 2.7290 | 0.0000 | |
| SRP064894 | PGF | 5228 | RNAseq | 2.1382 | 0.0000 | |
| SRP133303 | PGF | 5228 | RNAseq | 1.9187 | 0.0000 | |
| SRP159526 | PGF | 5228 | RNAseq | 0.9770 | 0.0179 | |
| SRP219564 | PGF | 5228 | RNAseq | 0.8801 | 0.0483 | |
| TCGA | PGF | 5228 | RNAseq | 1.1364 | 0.0000 |
Upregulated datasets: 7; Downregulated datasets: 0.
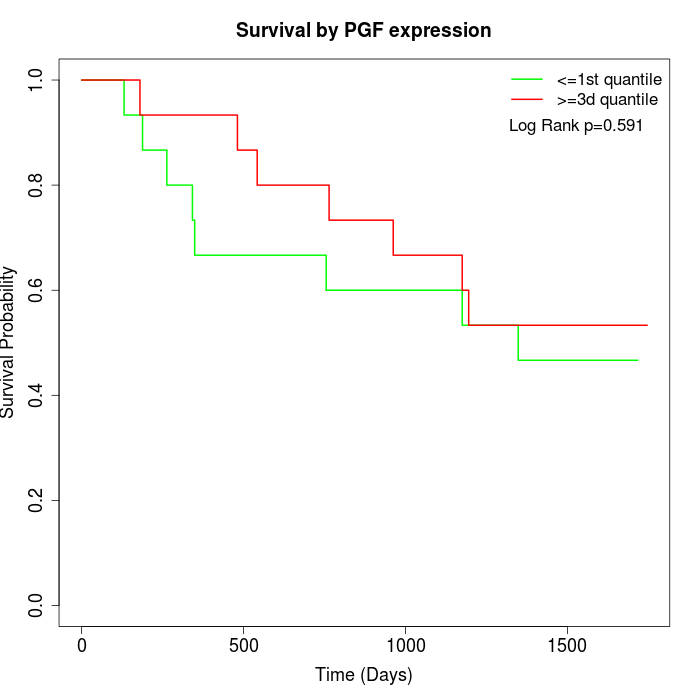
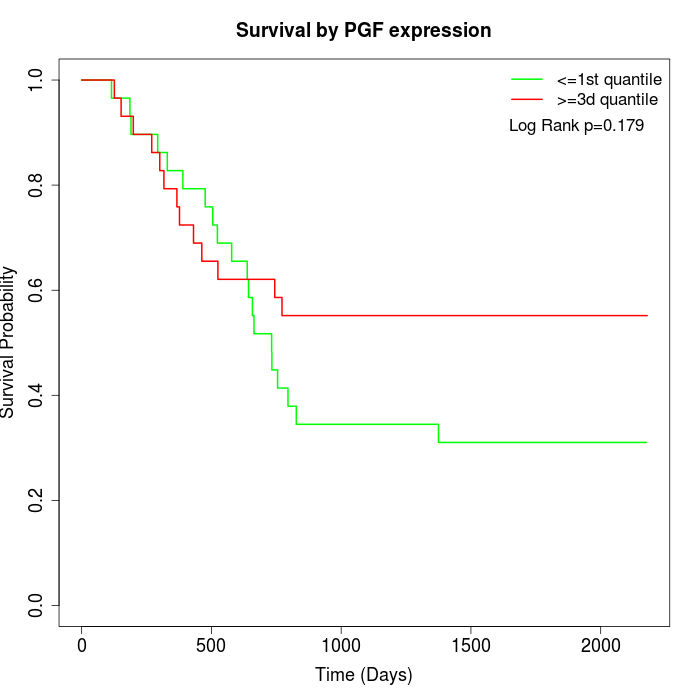
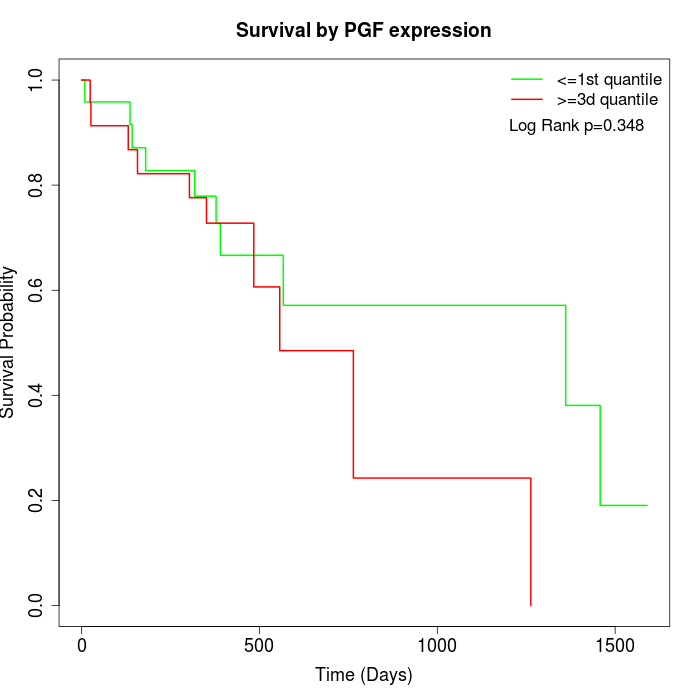
Survival by PGF expression:
Note: Click image to view full size file.
Copy number change of PGF:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PGF | 5228 | 8 | 3 | 19 | |
| GSE20123 | PGF | 5228 | 8 | 3 | 19 | |
| GSE43470 | PGF | 5228 | 8 | 3 | 32 | |
| GSE46452 | PGF | 5228 | 16 | 3 | 40 | |
| GSE47630 | PGF | 5228 | 11 | 8 | 21 | |
| GSE54993 | PGF | 5228 | 3 | 8 | 59 | |
| GSE54994 | PGF | 5228 | 19 | 4 | 30 | |
| GSE60625 | PGF | 5228 | 0 | 2 | 9 | |
| GSE74703 | PGF | 5228 | 7 | 3 | 26 | |
| GSE74704 | PGF | 5228 | 3 | 3 | 14 | |
| TCGA | PGF | 5228 | 32 | 16 | 48 |
Total number of gains: 115; Total number of losses: 56; Total Number of normals: 317.
Somatic mutations of PGF:
Generating mutation plots.
Highly correlated genes for PGF:
Showing top 20/206 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PGF | ZNF160 | 0.946476 | 9 | 0 | 9 |
| PGF | FBXW12 | 0.922101 | 9 | 0 | 9 |
| PGF | SLC22A3 | 0.908673 | 5 | 0 | 5 |
| PGF | CMBL | 0.894602 | 5 | 0 | 5 |
| PGF | ZNF611 | 0.891199 | 9 | 0 | 9 |
| PGF | CCDC152 | 0.868819 | 6 | 0 | 6 |
| PGF | TSR1 | 0.859035 | 8 | 0 | 7 |
| PGF | CDK13 | 0.856008 | 7 | 0 | 7 |
| PGF | FUT8-AS1 | 0.830746 | 4 | 0 | 4 |
| PGF | UACA | 0.80505 | 4 | 0 | 4 |
| PGF | ANKRD10-IT1 | 0.804641 | 9 | 0 | 8 |
| PGF | WDPCP | 0.778104 | 6 | 0 | 6 |
| PGF | TNRC6C-AS1 | 0.75837 | 5 | 0 | 5 |
| PGF | FPR1 | 0.750611 | 3 | 0 | 3 |
| PGF | C1QTNF6 | 0.746523 | 3 | 0 | 3 |
| PGF | SSBP3-AS1 | 0.745673 | 5 | 0 | 5 |
| PGF | XRCC2 | 0.744408 | 10 | 0 | 9 |
| PGF | ARHGEF1 | 0.738585 | 8 | 0 | 8 |
| PGF | CCDC142 | 0.727658 | 3 | 0 | 3 |
| PGF | GTSE1 | 0.716172 | 10 | 0 | 10 |
For details and further investigation, click here