| Full name: prolactin induced protein | Alias Symbol: GCDFP-15|GCDFP15|GPIP4 | ||
| Type: protein-coding gene | Cytoband: 7q34 | ||
| Entrez ID: 5304 | HGNC ID: HGNC:8993 | Ensembl Gene: ENSG00000159763 | OMIM ID: 176720 |
| Drug and gene relationship at DGIdb | |||
Expression of PIP:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PIP | 5304 | 206509_at | -0.3010 | 0.3326 | |
| GSE20347 | PIP | 5304 | 206509_at | -0.0570 | 0.5054 | |
| GSE23400 | PIP | 5304 | 206509_at | -0.1283 | 0.0002 | |
| GSE26886 | PIP | 5304 | 206509_at | -0.0470 | 0.7394 | |
| GSE29001 | PIP | 5304 | 206509_at | -0.0832 | 0.6505 | |
| GSE38129 | PIP | 5304 | 206509_at | -0.2401 | 0.0044 | |
| GSE45670 | PIP | 5304 | 206509_at | -0.2695 | 0.0505 | |
| GSE53622 | PIP | 5304 | 52266 | -0.4459 | 0.0421 | |
| GSE53624 | PIP | 5304 | 52266 | -1.0644 | 0.0000 | |
| GSE63941 | PIP | 5304 | 206509_at | 0.1692 | 0.3663 | |
| GSE77861 | PIP | 5304 | 206509_at | -0.1344 | 0.2146 | |
| TCGA | PIP | 5304 | RNAseq | 1.8160 | 0.3531 |
Upregulated datasets: 0; Downregulated datasets: 1.
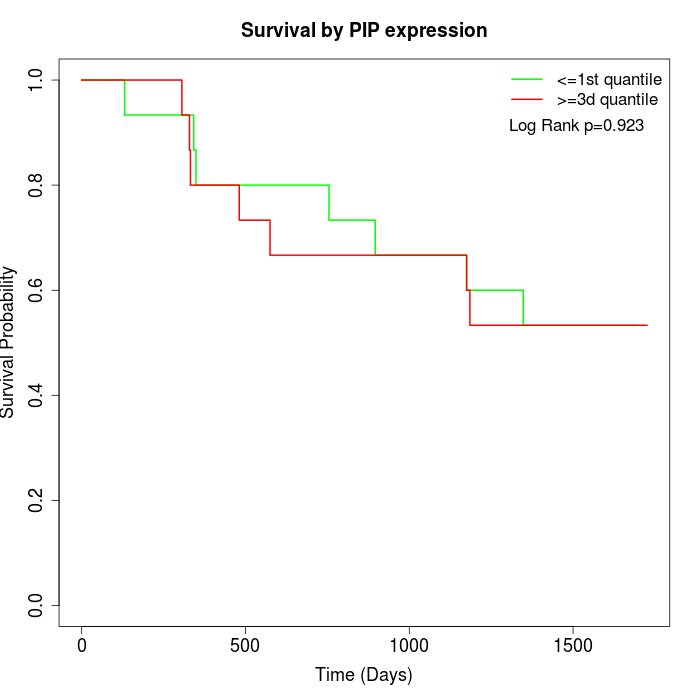
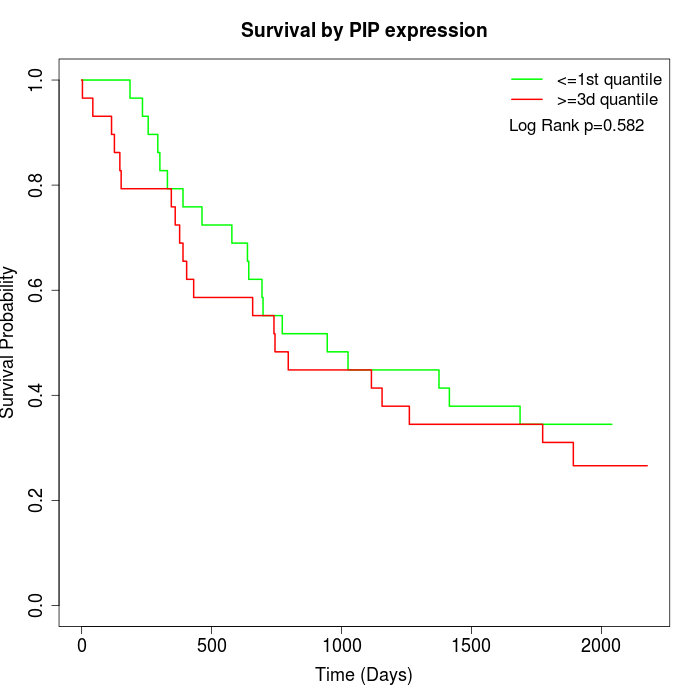
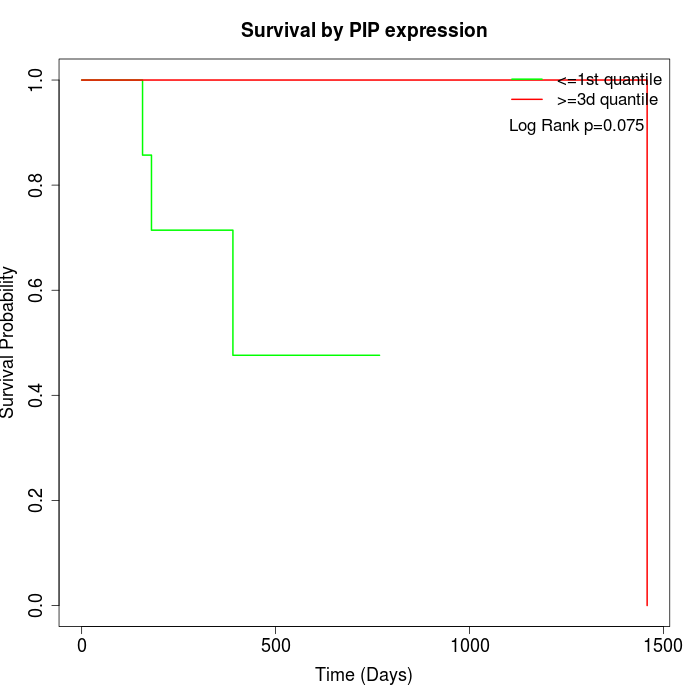
Survival by PIP expression:
Note: Click image to view full size file.
Copy number change of PIP:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PIP | 5304 | 4 | 2 | 24 | |
| GSE20123 | PIP | 5304 | 4 | 2 | 24 | |
| GSE43470 | PIP | 5304 | 2 | 4 | 37 | |
| GSE46452 | PIP | 5304 | 7 | 2 | 50 | |
| GSE47630 | PIP | 5304 | 6 | 7 | 27 | |
| GSE54993 | PIP | 5304 | 3 | 4 | 63 | |
| GSE54994 | PIP | 5304 | 5 | 8 | 40 | |
| GSE60625 | PIP | 5304 | 0 | 0 | 11 | |
| GSE74703 | PIP | 5304 | 2 | 4 | 30 | |
| GSE74704 | PIP | 5304 | 2 | 2 | 16 | |
| TCGA | PIP | 5304 | 28 | 24 | 44 |
Total number of gains: 63; Total number of losses: 59; Total Number of normals: 366.
Somatic mutations of PIP:
Generating mutation plots.
Highly correlated genes for PIP:
Showing top 20/546 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PIP | ZG16B | 0.752395 | 4 | 0 | 4 |
| PIP | BPIFB1 | 0.736669 | 4 | 0 | 4 |
| PIP | LCN1 | 0.723401 | 6 | 0 | 6 |
| PIP | C6orf58 | 0.71677 | 5 | 0 | 4 |
| PIP | AZGP1 | 0.710618 | 5 | 0 | 5 |
| PIP | BPIFB2 | 0.705505 | 6 | 0 | 6 |
| PIP | REG3A | 0.701698 | 3 | 0 | 3 |
| PIP | SSTR5-AS1 | 0.694821 | 3 | 0 | 3 |
| PIP | SCGB3A1 | 0.69356 | 5 | 0 | 4 |
| PIP | SCGB1D2 | 0.687539 | 8 | 0 | 7 |
| PIP | NDOR1 | 0.685667 | 4 | 0 | 4 |
| PIP | ADRB3 | 0.680164 | 4 | 0 | 4 |
| PIP | DIO1 | 0.676407 | 3 | 0 | 3 |
| PIP | CHRM1 | 0.673104 | 4 | 0 | 3 |
| PIP | PLIN1 | 0.671724 | 5 | 0 | 4 |
| PIP | EDA2R | 0.671139 | 4 | 0 | 4 |
| PIP | CHST4 | 0.67113 | 8 | 0 | 8 |
| PIP | IGH | 0.660552 | 3 | 0 | 3 |
| PIP | MUC7 | 0.65821 | 7 | 0 | 5 |
| PIP | ASCL3 | 0.656876 | 6 | 0 | 5 |
For details and further investigation, click here