| Full name: cholinergic receptor muscarinic 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 11q12.3 | ||
| Entrez ID: 1128 | HGNC ID: HGNC:1950 | Ensembl Gene: ENSG00000168539 | OMIM ID: 118510 |
| Drug and gene relationship at DGIdb | |||
CHRM1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04020 | Calcium signaling pathway | |
| hsa04024 | cAMP signaling pathway | |
| hsa04151 | PI3K-Akt signaling pathway | |
| hsa04725 | Cholinergic synapse | |
| hsa04810 | Regulation of actin cytoskeleton |
Expression of CHRM1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CHRM1 | 1128 | 231783_at | -0.2868 | 0.2421 | |
| GSE26886 | CHRM1 | 1128 | 231783_at | 0.0270 | 0.8533 | |
| GSE45670 | CHRM1 | 1128 | 231783_at | -0.1029 | 0.2676 | |
| GSE53622 | CHRM1 | 1128 | 138271 | -1.4584 | 0.0000 | |
| GSE53624 | CHRM1 | 1128 | 138271 | -1.6394 | 0.0000 | |
| GSE63941 | CHRM1 | 1128 | 231783_at | 0.0709 | 0.5929 | |
| GSE77861 | CHRM1 | 1128 | 231783_at | -0.1792 | 0.0410 | |
| TCGA | CHRM1 | 1128 | RNAseq | -1.8309 | 0.0073 |
Upregulated datasets: 0; Downregulated datasets: 3.
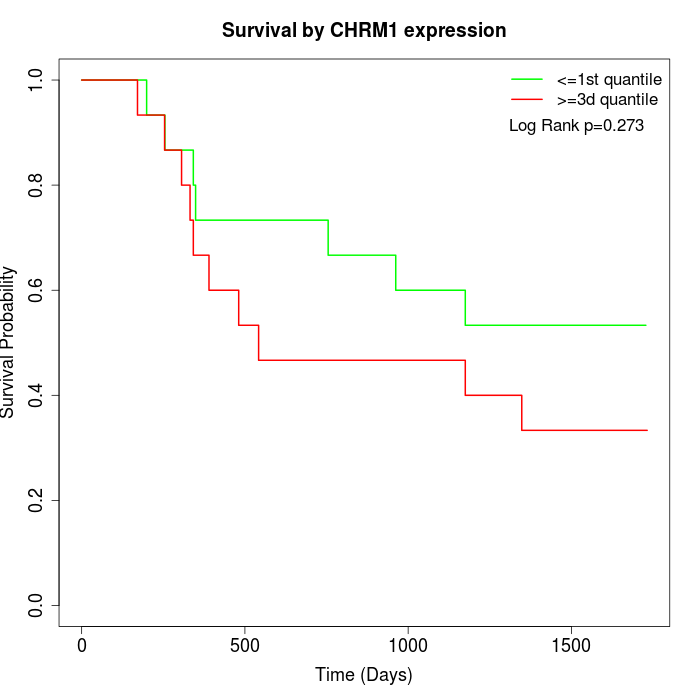
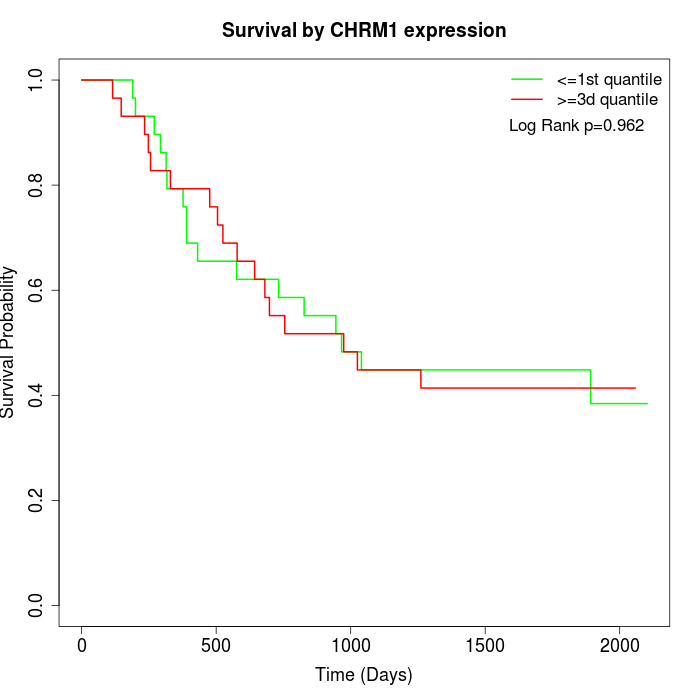
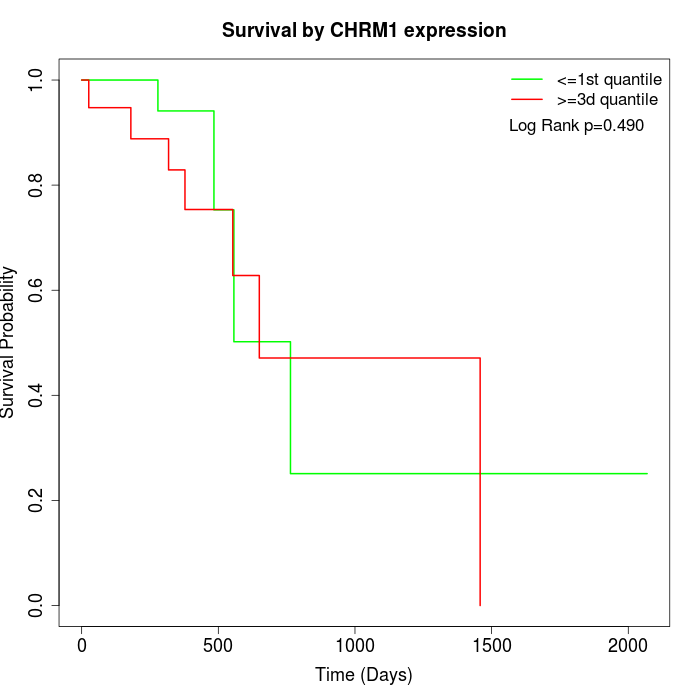
Survival by CHRM1 expression:
Note: Click image to view full size file.
Copy number change of CHRM1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CHRM1 | 1128 | 6 | 5 | 19 | |
| GSE20123 | CHRM1 | 1128 | 6 | 5 | 19 | |
| GSE43470 | CHRM1 | 1128 | 1 | 6 | 36 | |
| GSE46452 | CHRM1 | 1128 | 8 | 4 | 47 | |
| GSE47630 | CHRM1 | 1128 | 3 | 7 | 30 | |
| GSE54993 | CHRM1 | 1128 | 3 | 0 | 67 | |
| GSE54994 | CHRM1 | 1128 | 5 | 5 | 43 | |
| GSE60625 | CHRM1 | 1128 | 0 | 3 | 8 | |
| GSE74703 | CHRM1 | 1128 | 1 | 3 | 32 | |
| GSE74704 | CHRM1 | 1128 | 4 | 3 | 13 | |
| TCGA | CHRM1 | 1128 | 16 | 11 | 69 |
Total number of gains: 53; Total number of losses: 52; Total Number of normals: 383.
Somatic mutations of CHRM1:
Generating mutation plots.
Highly correlated genes for CHRM1:
Showing top 20/99 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CHRM1 | NXPE4 | 0.744052 | 4 | 0 | 4 |
| CHRM1 | SMC5-AS1 | 0.741077 | 3 | 0 | 3 |
| CHRM1 | C6orf58 | 0.740988 | 4 | 0 | 4 |
| CHRM1 | SSTR5-AS1 | 0.738105 | 3 | 0 | 3 |
| CHRM1 | NKX3-1 | 0.733522 | 4 | 0 | 4 |
| CHRM1 | TFF3 | 0.722454 | 5 | 0 | 4 |
| CHRM1 | TSPAN8 | 0.721005 | 5 | 0 | 5 |
| CHRM1 | AZGP1 | 0.708539 | 4 | 0 | 4 |
| CHRM1 | MS4A8 | 0.69661 | 3 | 0 | 3 |
| CHRM1 | BPIFB2 | 0.694687 | 5 | 0 | 4 |
| CHRM1 | TOX3 | 0.692907 | 3 | 0 | 3 |
| CHRM1 | SCGB1D1 | 0.692248 | 3 | 0 | 3 |
| CHRM1 | SCGB2A1 | 0.688164 | 3 | 0 | 3 |
| CHRM1 | SCGB2A2 | 0.681289 | 4 | 0 | 4 |
| CHRM1 | STATH | 0.676715 | 6 | 0 | 6 |
| CHRM1 | PIP | 0.673104 | 4 | 0 | 3 |
| CHRM1 | FCRL4 | 0.665985 | 3 | 0 | 3 |
| CHRM1 | SCGB1D2 | 0.663646 | 5 | 0 | 4 |
| CHRM1 | PPP1R1B | 0.661164 | 4 | 0 | 3 |
| CHRM1 | EHF | 0.655915 | 3 | 0 | 3 |
For details and further investigation, click here