| Full name: phosphoinositide interacting regulator of transient receptor potential channels | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 17p12 | ||
| Entrez ID: 644139 | HGNC ID: HGNC:37239 | Ensembl Gene: ENSG00000233670 | OMIM ID: 612068 |
| Drug and gene relationship at DGIdb | |||
Expression of PIRT:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PIRT | 644139 | 232887_at | -0.1669 | 0.6904 | |
| GSE26886 | PIRT | 644139 | 232887_at | 0.0271 | 0.8119 | |
| GSE45670 | PIRT | 644139 | 232887_at | 0.0865 | 0.4429 | |
| GSE53622 | PIRT | 644139 | 5221 | -0.2071 | 0.3880 | |
| GSE53624 | PIRT | 644139 | 5221 | -0.4764 | 0.0021 | |
| GSE63941 | PIRT | 644139 | 232887_at | 0.1827 | 0.2833 | |
| GSE77861 | PIRT | 644139 | 232887_at | -0.1048 | 0.3122 | |
| TCGA | PIRT | 644139 | RNAseq | -3.4496 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
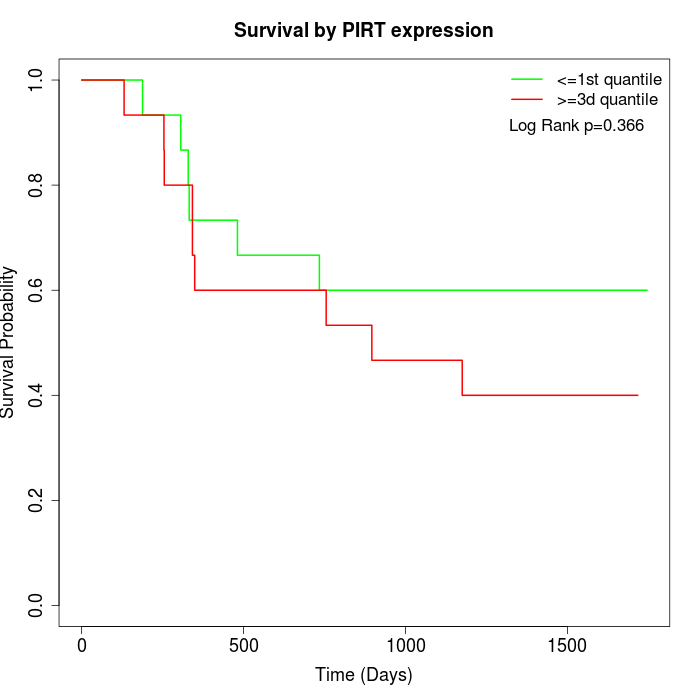
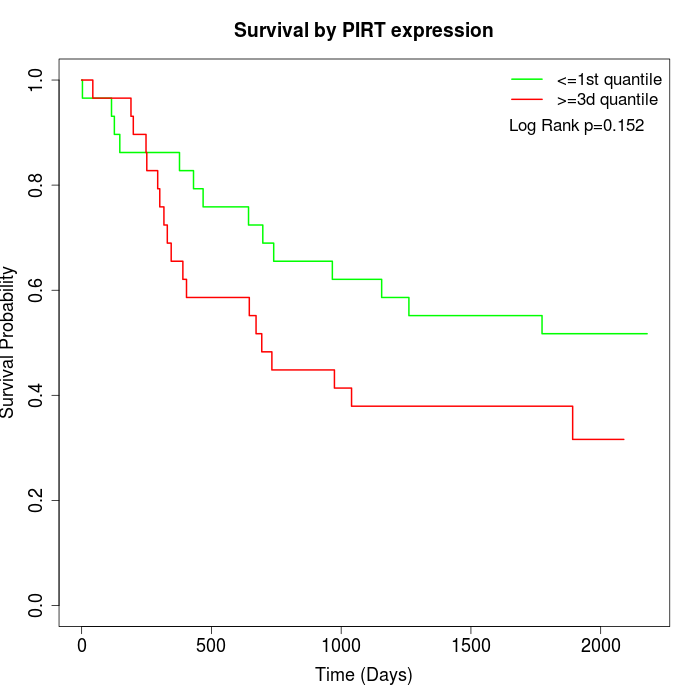
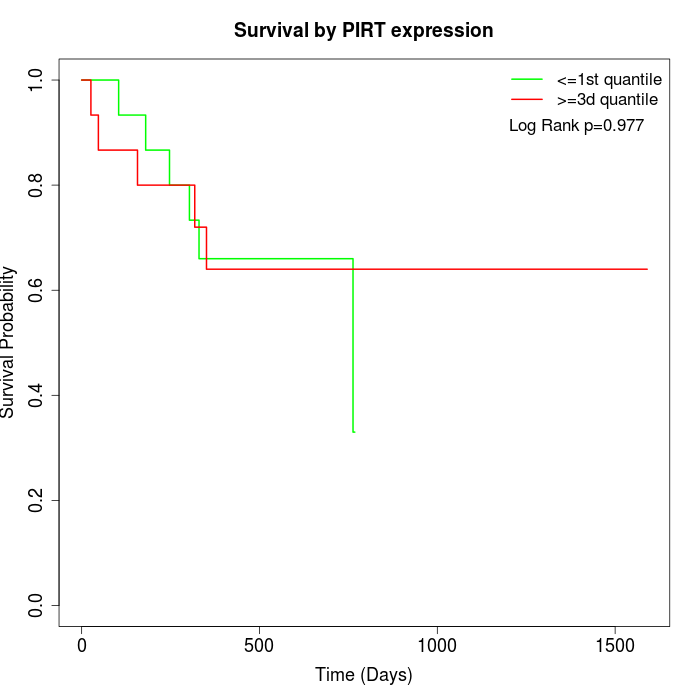
Survival by PIRT expression:
Note: Click image to view full size file.
Copy number change of PIRT:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PIRT | 644139 | 3 | 4 | 23 | |
| GSE20123 | PIRT | 644139 | 3 | 5 | 22 | |
| GSE43470 | PIRT | 644139 | 2 | 5 | 36 | |
| GSE46452 | PIRT | 644139 | 34 | 1 | 24 | |
| GSE47630 | PIRT | 644139 | 7 | 1 | 32 | |
| GSE54993 | PIRT | 644139 | 3 | 3 | 64 | |
| GSE54994 | PIRT | 644139 | 5 | 7 | 41 | |
| GSE60625 | PIRT | 644139 | 4 | 0 | 7 | |
| GSE74703 | PIRT | 644139 | 2 | 3 | 31 | |
| GSE74704 | PIRT | 644139 | 2 | 2 | 16 | |
| TCGA | PIRT | 644139 | 18 | 20 | 58 |
Total number of gains: 83; Total number of losses: 51; Total Number of normals: 354.
Somatic mutations of PIRT:
Generating mutation plots.
Highly correlated genes for PIRT:
Showing top 20/24 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PIRT | SNAP91 | 0.727219 | 3 | 0 | 3 |
| PIRT | SLC35F1 | 0.707003 | 3 | 0 | 3 |
| PIRT | ZMAT4 | 0.686745 | 3 | 0 | 3 |
| PIRT | ARHGAP6 | 0.627342 | 3 | 0 | 3 |
| PIRT | LMO3 | 0.606493 | 3 | 0 | 3 |
| PIRT | RNF122 | 0.595779 | 3 | 0 | 3 |
| PIRT | NPY | 0.583537 | 4 | 0 | 3 |
| PIRT | LRRC4C | 0.580836 | 5 | 0 | 4 |
| PIRT | LINC00642 | 0.577354 | 3 | 0 | 3 |
| PIRT | PAGE4 | 0.573573 | 4 | 0 | 3 |
| PIRT | COL26A1 | 0.554058 | 3 | 0 | 3 |
| PIRT | CA14 | 0.549447 | 5 | 0 | 3 |
| PIRT | ENAM | 0.548694 | 4 | 0 | 3 |
| PIRT | MAP6 | 0.548179 | 4 | 0 | 3 |
| PIRT | TGM7 | 0.546359 | 5 | 0 | 3 |
| PIRT | STMN2 | 0.545263 | 6 | 0 | 3 |
| PIRT | CNTN2 | 0.538888 | 4 | 0 | 3 |
| PIRT | ZNF169 | 0.538219 | 4 | 0 | 3 |
| PIRT | SRRM4 | 0.526959 | 4 | 0 | 3 |
| PIRT | DRP2 | 0.51967 | 4 | 0 | 3 |
For details and further investigation, click here