| Full name: microtubule associated protein 6 | Alias Symbol: KIAA1878|STOP|FLJ41346|MAP6-N | ||
| Type: protein-coding gene | Cytoband: 11q13.5 | ||
| Entrez ID: 4135 | HGNC ID: HGNC:6868 | Ensembl Gene: ENSG00000171533 | OMIM ID: 601783 |
| Drug and gene relationship at DGIdb | |||
Expression of MAP6:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | MAP6 | 4135 | 228943_at | -0.5831 | 0.1538 | |
| GSE26886 | MAP6 | 4135 | 228943_at | -0.0181 | 0.9224 | |
| GSE45670 | MAP6 | 4135 | 228943_at | -0.5454 | 0.0001 | |
| GSE53622 | MAP6 | 4135 | 138362 | -0.3619 | 0.0004 | |
| GSE53624 | MAP6 | 4135 | 138362 | -0.4623 | 0.0000 | |
| GSE63941 | MAP6 | 4135 | 228943_at | -0.3440 | 0.1242 | |
| GSE77861 | MAP6 | 4135 | 228943_at | -0.2797 | 0.0110 | |
| GSE97050 | MAP6 | 4135 | A_23_P47728 | -1.5186 | 0.0620 | |
| SRP064894 | MAP6 | 4135 | RNAseq | 0.2587 | 0.5608 | |
| SRP133303 | MAP6 | 4135 | RNAseq | -1.6164 | 0.0000 | |
| SRP159526 | MAP6 | 4135 | RNAseq | 0.1789 | 0.6241 | |
| SRP219564 | MAP6 | 4135 | RNAseq | -0.9515 | 0.3728 | |
| TCGA | MAP6 | 4135 | RNAseq | -0.9723 | 0.0007 |
Upregulated datasets: 0; Downregulated datasets: 1.
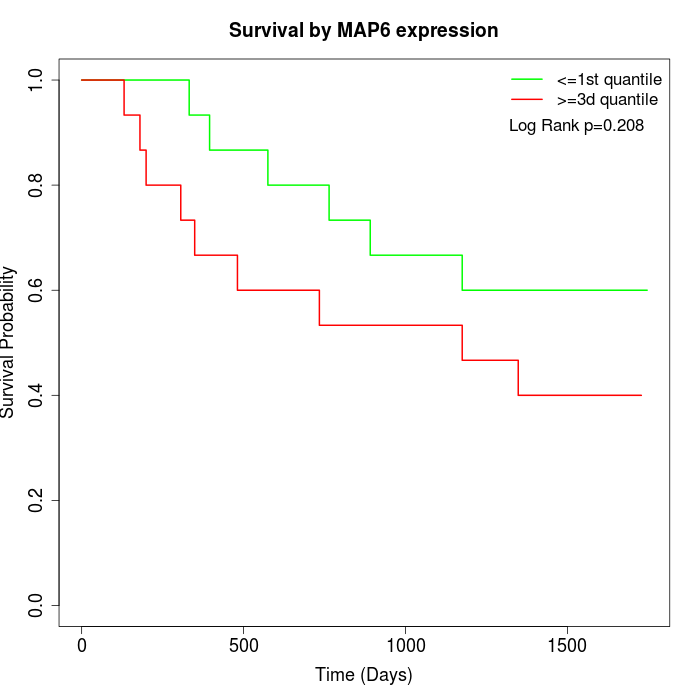
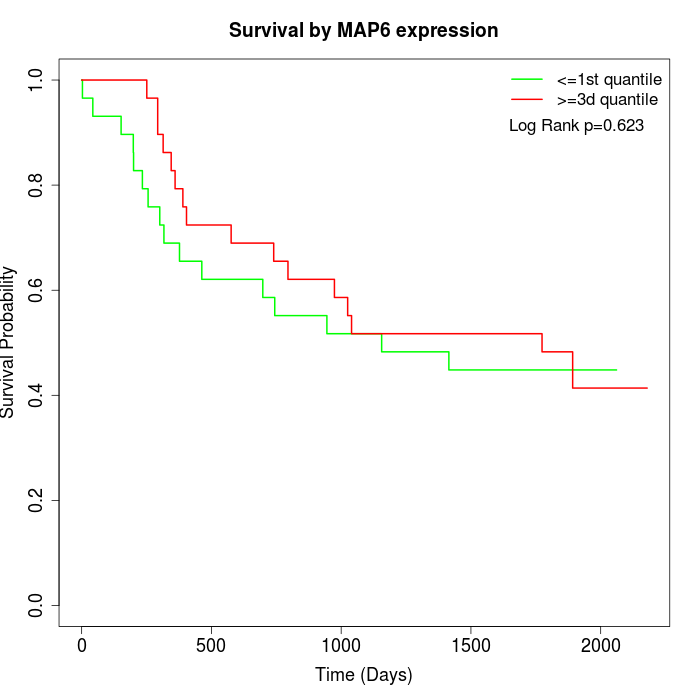
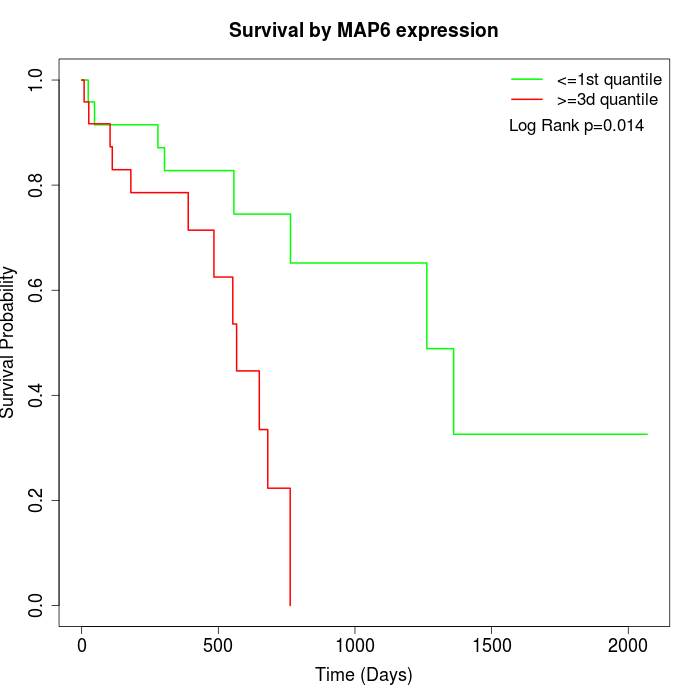
Survival by MAP6 expression:
Note: Click image to view full size file.
Copy number change of MAP6:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | MAP6 | 4135 | 6 | 8 | 16 | |
| GSE20123 | MAP6 | 4135 | 6 | 7 | 17 | |
| GSE43470 | MAP6 | 4135 | 4 | 4 | 35 | |
| GSE46452 | MAP6 | 4135 | 7 | 22 | 30 | |
| GSE47630 | MAP6 | 4135 | 6 | 14 | 20 | |
| GSE54993 | MAP6 | 4135 | 8 | 2 | 60 | |
| GSE54994 | MAP6 | 4135 | 7 | 16 | 30 | |
| GSE60625 | MAP6 | 4135 | 4 | 3 | 4 | |
| GSE74703 | MAP6 | 4135 | 3 | 3 | 30 | |
| GSE74704 | MAP6 | 4135 | 4 | 3 | 13 | |
| TCGA | MAP6 | 4135 | 21 | 31 | 44 |
Total number of gains: 76; Total number of losses: 113; Total Number of normals: 299.
Somatic mutations of MAP6:
Generating mutation plots.
Highly correlated genes for MAP6:
Showing top 20/501 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| MAP6 | PPP1R12B | 0.718805 | 4 | 0 | 4 |
| MAP6 | TCEAL2 | 0.718383 | 5 | 0 | 5 |
| MAP6 | RYR2 | 0.716834 | 4 | 0 | 4 |
| MAP6 | ZNF662 | 0.710443 | 3 | 0 | 3 |
| MAP6 | DES | 0.705835 | 4 | 0 | 3 |
| MAP6 | SLC4A4 | 0.704987 | 4 | 0 | 4 |
| MAP6 | TFAP2B | 0.704019 | 3 | 0 | 3 |
| MAP6 | DDAH1 | 0.703002 | 3 | 0 | 3 |
| MAP6 | SH3BGRL2 | 0.702558 | 3 | 0 | 3 |
| MAP6 | FIG4 | 0.699871 | 3 | 0 | 3 |
| MAP6 | FENDRR | 0.698837 | 4 | 0 | 4 |
| MAP6 | SORT1 | 0.691757 | 3 | 0 | 3 |
| MAP6 | FAM13B | 0.689965 | 3 | 0 | 3 |
| MAP6 | VIT | 0.688401 | 4 | 0 | 3 |
| MAP6 | RAI2 | 0.688394 | 4 | 0 | 4 |
| MAP6 | GYPC | 0.68674 | 5 | 0 | 4 |
| MAP6 | DCLK2 | 0.683322 | 3 | 0 | 3 |
| MAP6 | KCNMB1 | 0.683265 | 4 | 0 | 3 |
| MAP6 | HDAC5 | 0.680207 | 3 | 0 | 3 |
| MAP6 | AK9 | 0.677624 | 4 | 0 | 3 |
For details and further investigation, click here