| Full name: zinc finger matrin-type 4 | Alias Symbol: FLJ13842 | ||
| Type: protein-coding gene | Cytoband: 8p11.21 | ||
| Entrez ID: 79698 | HGNC ID: HGNC:25844 | Ensembl Gene: ENSG00000165061 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of ZMAT4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ZMAT4 | 79698 | 219877_at | -0.2857 | 0.5803 | |
| GSE20347 | ZMAT4 | 79698 | 219877_at | 0.1373 | 0.2585 | |
| GSE23400 | ZMAT4 | 79698 | 219877_at | -0.3002 | 0.0000 | |
| GSE26886 | ZMAT4 | 79698 | 219877_at | 0.2823 | 0.0592 | |
| GSE29001 | ZMAT4 | 79698 | 219877_at | -0.1232 | 0.5112 | |
| GSE38129 | ZMAT4 | 79698 | 219877_at | 0.0106 | 0.9524 | |
| GSE45670 | ZMAT4 | 79698 | 219877_at | 0.0261 | 0.8849 | |
| GSE53622 | ZMAT4 | 79698 | 18528 | 0.4853 | 0.0917 | |
| GSE53624 | ZMAT4 | 79698 | 165523 | 0.8561 | 0.0000 | |
| GSE63941 | ZMAT4 | 79698 | 219877_at | -0.1625 | 0.6692 | |
| GSE77861 | ZMAT4 | 79698 | 219877_at | -0.1415 | 0.1842 | |
| GSE97050 | ZMAT4 | 79698 | A_33_P3216133 | -0.7409 | 0.2987 | |
| TCGA | ZMAT4 | 79698 | RNAseq | 1.1925 | 0.3179 |
Upregulated datasets: 0; Downregulated datasets: 0.
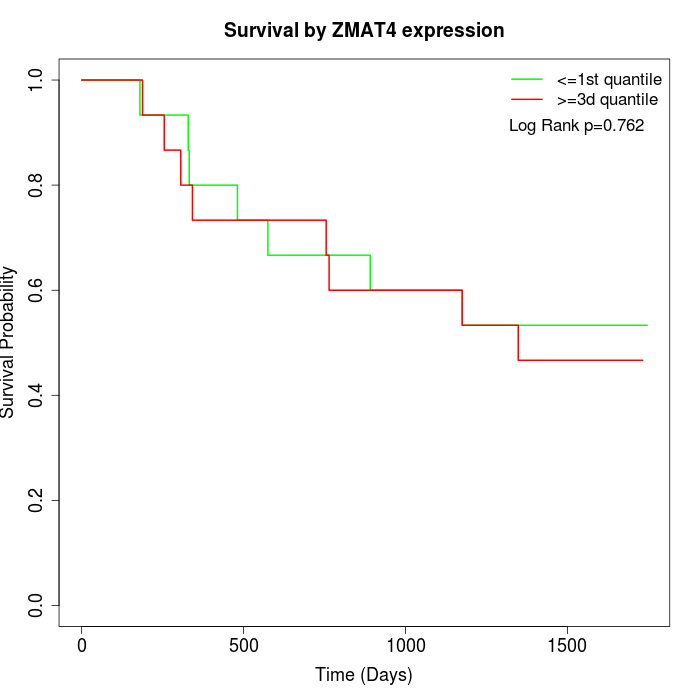
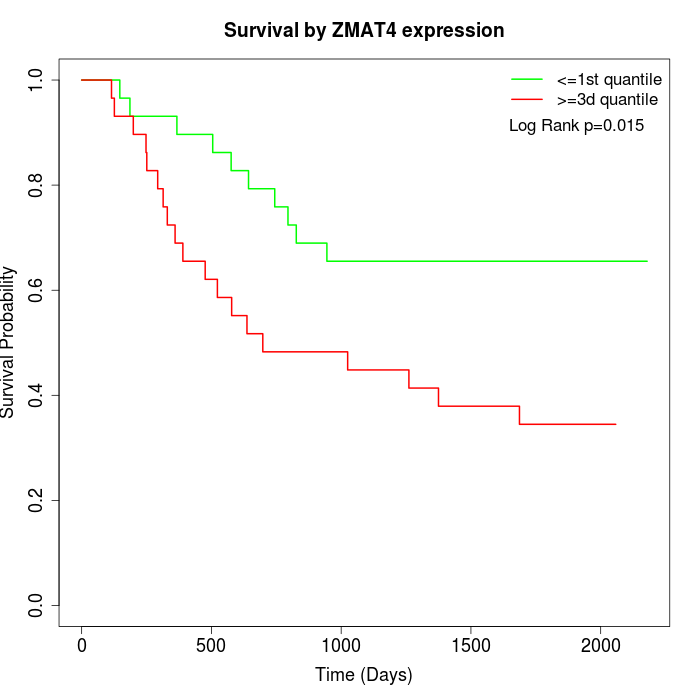
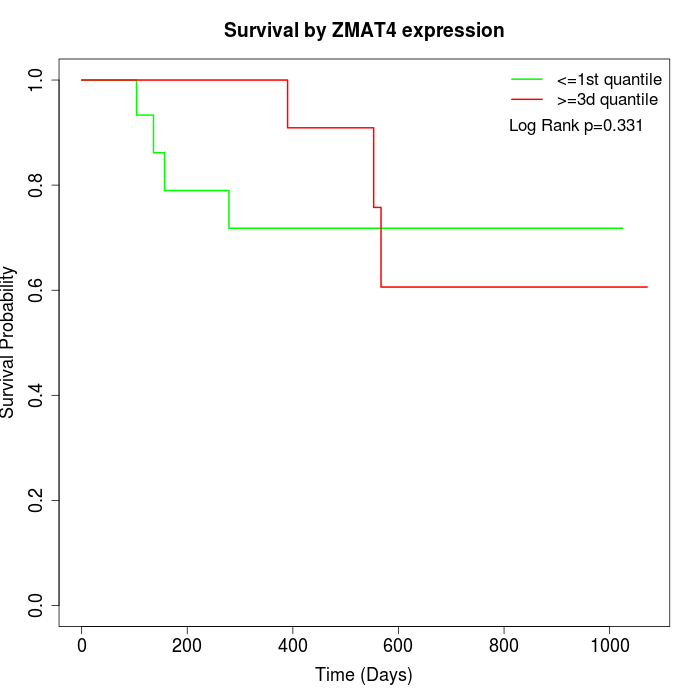
Survival by ZMAT4 expression:
Note: Click image to view full size file.
Copy number change of ZMAT4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ZMAT4 | 79698 | 8 | 5 | 17 | |
| GSE20123 | ZMAT4 | 79698 | 8 | 5 | 17 | |
| GSE43470 | ZMAT4 | 79698 | 7 | 6 | 30 | |
| GSE46452 | ZMAT4 | 79698 | 20 | 5 | 34 | |
| GSE47630 | ZMAT4 | 79698 | 22 | 1 | 17 | |
| GSE54993 | ZMAT4 | 79698 | 2 | 17 | 51 | |
| GSE54994 | ZMAT4 | 79698 | 19 | 8 | 26 | |
| GSE60625 | ZMAT4 | 79698 | 3 | 0 | 8 | |
| GSE74703 | ZMAT4 | 79698 | 7 | 5 | 24 | |
| GSE74704 | ZMAT4 | 79698 | 7 | 2 | 11 | |
| TCGA | ZMAT4 | 79698 | 36 | 22 | 38 |
Total number of gains: 139; Total number of losses: 76; Total Number of normals: 273.
Somatic mutations of ZMAT4:
Generating mutation plots.
Highly correlated genes for ZMAT4:
Showing top 20/882 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ZMAT4 | MYEF2 | 0.78065 | 5 | 0 | 5 |
| ZMAT4 | LUZP4 | 0.77335 | 4 | 0 | 4 |
| ZMAT4 | AKAP6 | 0.772466 | 5 | 0 | 5 |
| ZMAT4 | ADAL | 0.770285 | 3 | 0 | 3 |
| ZMAT4 | CGA | 0.762102 | 3 | 0 | 3 |
| ZMAT4 | CTNNA2 | 0.7612 | 3 | 0 | 3 |
| ZMAT4 | KIR2DS5 | 0.760663 | 4 | 0 | 4 |
| ZMAT4 | RCVRN | 0.759877 | 4 | 0 | 4 |
| ZMAT4 | MTNR1B | 0.759328 | 3 | 0 | 3 |
| ZMAT4 | GALNT8 | 0.75861 | 3 | 0 | 3 |
| ZMAT4 | RPGRIP1 | 0.755747 | 3 | 0 | 3 |
| ZMAT4 | HAPLN1 | 0.755216 | 3 | 0 | 3 |
| ZMAT4 | ART1 | 0.747985 | 3 | 0 | 3 |
| ZMAT4 | NPTX1 | 0.74436 | 5 | 0 | 5 |
| ZMAT4 | KCNJ5 | 0.743493 | 3 | 0 | 3 |
| ZMAT4 | CAPN11 | 0.73955 | 3 | 0 | 3 |
| ZMAT4 | KCNA1 | 0.73929 | 3 | 0 | 3 |
| ZMAT4 | ATN1 | 0.736513 | 3 | 0 | 3 |
| ZMAT4 | CYP19A1 | 0.736114 | 3 | 0 | 3 |
| ZMAT4 | RPH3A | 0.733706 | 3 | 0 | 3 |
For details and further investigation, click here