| Full name: phospholipase C eta 2 | Alias Symbol: KIAA0450|PLCeta2|RP3-395M20.1|PLC-eta2 | ||
| Type: protein-coding gene | Cytoband: 1p36.32 | ||
| Entrez ID: 9651 | HGNC ID: HGNC:29037 | Ensembl Gene: ENSG00000149527 | OMIM ID: 612836 |
| Drug and gene relationship at DGIdb | |||
Expression of PLCH2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PLCH2 | 9651 | 206080_at | -0.0047 | 0.9948 | |
| GSE20347 | PLCH2 | 9651 | 206080_at | -0.0951 | 0.4596 | |
| GSE23400 | PLCH2 | 9651 | 206080_at | -0.0169 | 0.8068 | |
| GSE26886 | PLCH2 | 9651 | 206080_at | 0.2101 | 0.2169 | |
| GSE29001 | PLCH2 | 9651 | 206080_at | -0.4078 | 0.0719 | |
| GSE38129 | PLCH2 | 9651 | 206080_at | -0.0420 | 0.8233 | |
| GSE45670 | PLCH2 | 9651 | 206080_at | 0.1324 | 0.2655 | |
| GSE53622 | PLCH2 | 9651 | 47488 | -0.2210 | 0.0013 | |
| GSE53624 | PLCH2 | 9651 | 47488 | -0.2597 | 0.0010 | |
| GSE63941 | PLCH2 | 9651 | 206080_at | 0.2946 | 0.2108 | |
| GSE77861 | PLCH2 | 9651 | 206080_at | -0.1616 | 0.1563 | |
| GSE97050 | PLCH2 | 9651 | A_24_P21887 | 0.0155 | 0.9611 | |
| SRP007169 | PLCH2 | 9651 | RNAseq | -0.1504 | 0.8123 | |
| SRP064894 | PLCH2 | 9651 | RNAseq | -0.4429 | 0.3139 | |
| SRP133303 | PLCH2 | 9651 | RNAseq | -0.6324 | 0.0110 | |
| SRP159526 | PLCH2 | 9651 | RNAseq | -1.7094 | 0.0016 | |
| SRP193095 | PLCH2 | 9651 | RNAseq | -0.0713 | 0.8224 | |
| SRP219564 | PLCH2 | 9651 | RNAseq | 0.6381 | 0.5654 | |
| TCGA | PLCH2 | 9651 | RNAseq | 0.8890 | 0.0001 |
Upregulated datasets: 0; Downregulated datasets: 1.
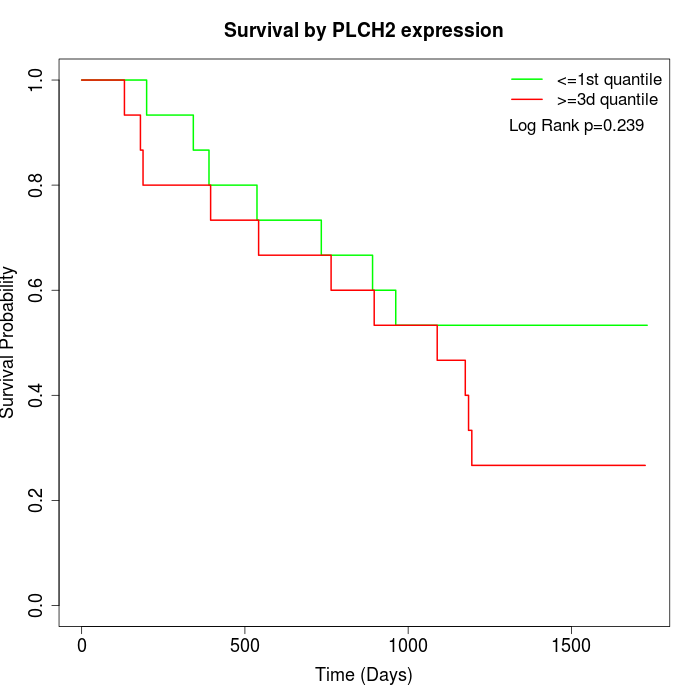
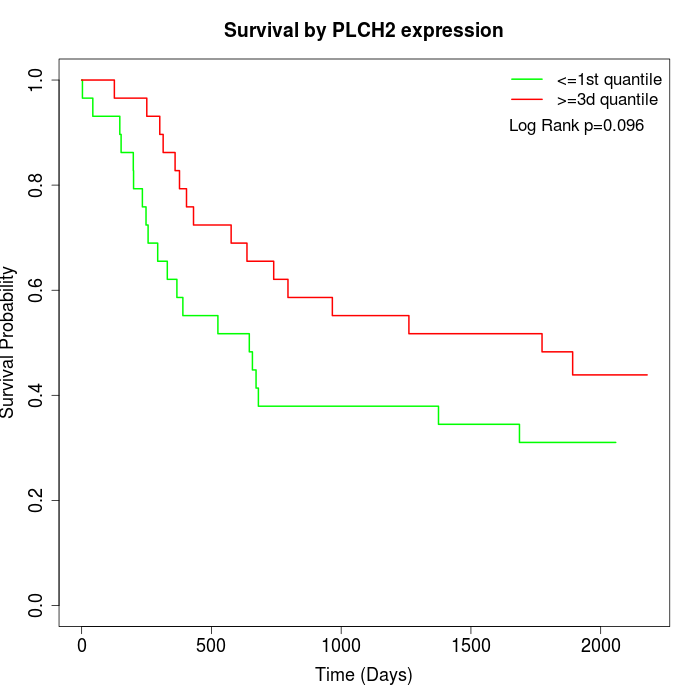
Survival by PLCH2 expression:
Note: Click image to view full size file.
Copy number change of PLCH2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PLCH2 | 9651 | 4 | 6 | 20 | |
| GSE20123 | PLCH2 | 9651 | 4 | 5 | 21 | |
| GSE43470 | PLCH2 | 9651 | 3 | 5 | 35 | |
| GSE46452 | PLCH2 | 9651 | 7 | 1 | 51 | |
| GSE47630 | PLCH2 | 9651 | 8 | 4 | 28 | |
| GSE54993 | PLCH2 | 9651 | 3 | 2 | 65 | |
| GSE54994 | PLCH2 | 9651 | 15 | 2 | 36 | |
| GSE60625 | PLCH2 | 9651 | 0 | 0 | 11 | |
| GSE74703 | PLCH2 | 9651 | 3 | 3 | 30 | |
| GSE74704 | PLCH2 | 9651 | 4 | 0 | 16 | |
| TCGA | PLCH2 | 9651 | 14 | 20 | 62 |
Total number of gains: 65; Total number of losses: 48; Total Number of normals: 375.
Somatic mutations of PLCH2:
Generating mutation plots.
Highly correlated genes for PLCH2:
Showing top 20/472 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PLCH2 | PTPRN | 0.71824 | 4 | 0 | 4 |
| PLCH2 | KBTBD13 | 0.712426 | 3 | 0 | 3 |
| PLCH2 | KIF19 | 0.711733 | 3 | 0 | 3 |
| PLCH2 | CLRN2 | 0.696966 | 3 | 0 | 3 |
| PLCH2 | RUFY4 | 0.695697 | 3 | 0 | 3 |
| PLCH2 | KRTAP6-1 | 0.693544 | 3 | 0 | 3 |
| PLCH2 | C19orf81 | 0.692043 | 4 | 0 | 3 |
| PLCH2 | PGC | 0.691705 | 4 | 0 | 3 |
| PLCH2 | MUC8 | 0.691506 | 4 | 0 | 3 |
| PLCH2 | CYP2D6 | 0.690422 | 5 | 0 | 4 |
| PLCH2 | C19orf44 | 0.689587 | 3 | 0 | 3 |
| PLCH2 | NAT8B | 0.686684 | 3 | 0 | 3 |
| PLCH2 | CD164L2 | 0.685178 | 3 | 0 | 3 |
| PLCH2 | TPRX1 | 0.677193 | 3 | 0 | 3 |
| PLCH2 | CCDC57 | 0.670699 | 4 | 0 | 4 |
| PLCH2 | TMEM212 | 0.67062 | 3 | 0 | 3 |
| PLCH2 | HHATL | 0.667773 | 4 | 0 | 4 |
| PLCH2 | OR13G1 | 0.660246 | 3 | 0 | 3 |
| PLCH2 | TM6SF2 | 0.658203 | 4 | 0 | 3 |
| PLCH2 | IPO13 | 0.656646 | 4 | 0 | 4 |
For details and further investigation, click here