| Full name: protein kinase cAMP-activated catalytic subunit gamma | Alias Symbol: PKACg | ||
| Type: protein-coding gene | Cytoband: 9q21.11 | ||
| Entrez ID: 5568 | HGNC ID: HGNC:9382 | Ensembl Gene: ENSG00000165059 | OMIM ID: 176893 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
PRKACG involved pathways:
Expression of PRKACG:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PRKACG | 5568 | 207228_at | -0.0621 | 0.8149 | |
| GSE20347 | PRKACG | 5568 | 207228_at | 0.0194 | 0.8222 | |
| GSE23400 | PRKACG | 5568 | 207228_at | -0.0613 | 0.0424 | |
| GSE26886 | PRKACG | 5568 | 207228_at | -0.1379 | 0.1187 | |
| GSE29001 | PRKACG | 5568 | 207228_at | -0.1221 | 0.3336 | |
| GSE38129 | PRKACG | 5568 | 207228_at | -0.1169 | 0.1535 | |
| GSE45670 | PRKACG | 5568 | 207228_at | -0.0626 | 0.4331 | |
| GSE53622 | PRKACG | 5568 | 38632 | -0.1736 | 0.0481 | |
| GSE53624 | PRKACG | 5568 | 38632 | -0.0773 | 0.4106 | |
| GSE63941 | PRKACG | 5568 | 207228_at | 0.0342 | 0.8257 | |
| GSE77861 | PRKACG | 5568 | 207228_at | -0.2548 | 0.0203 | |
| GSE97050 | PRKACG | 5568 | A_23_P71926 | -0.0304 | 0.9133 |
Upregulated datasets: 0; Downregulated datasets: 0.
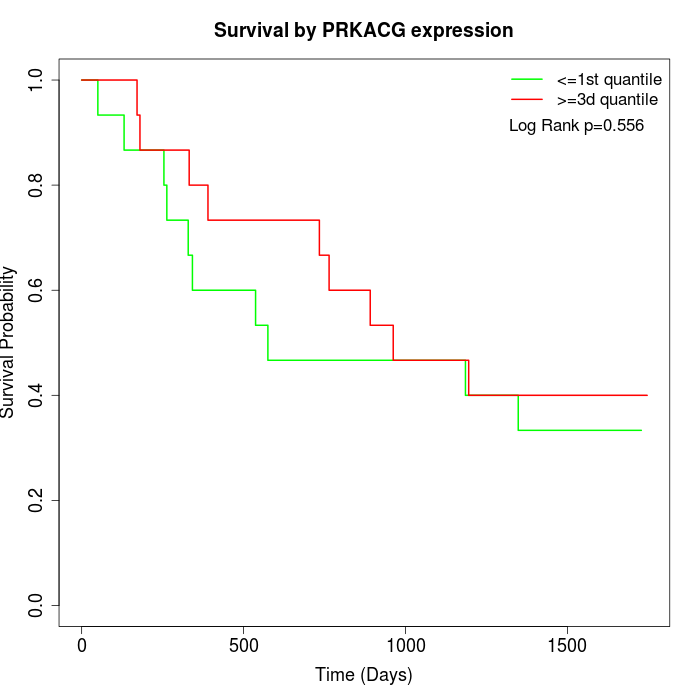
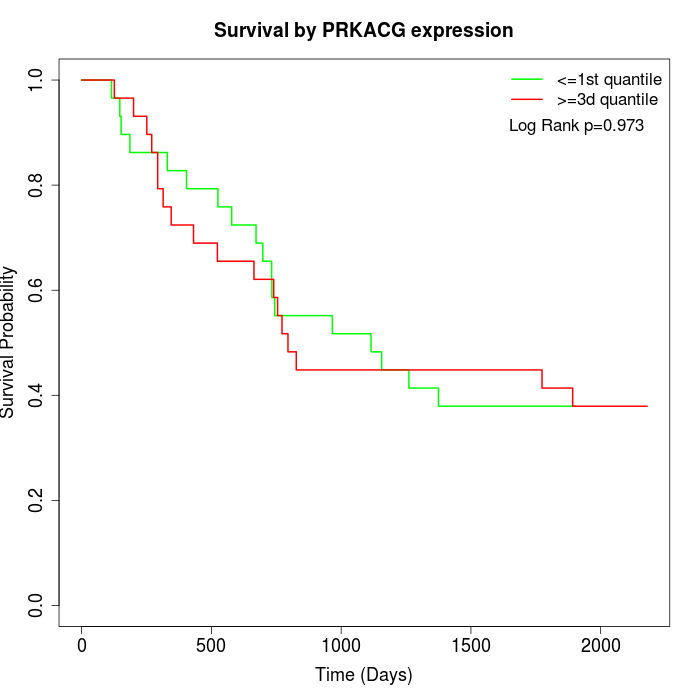
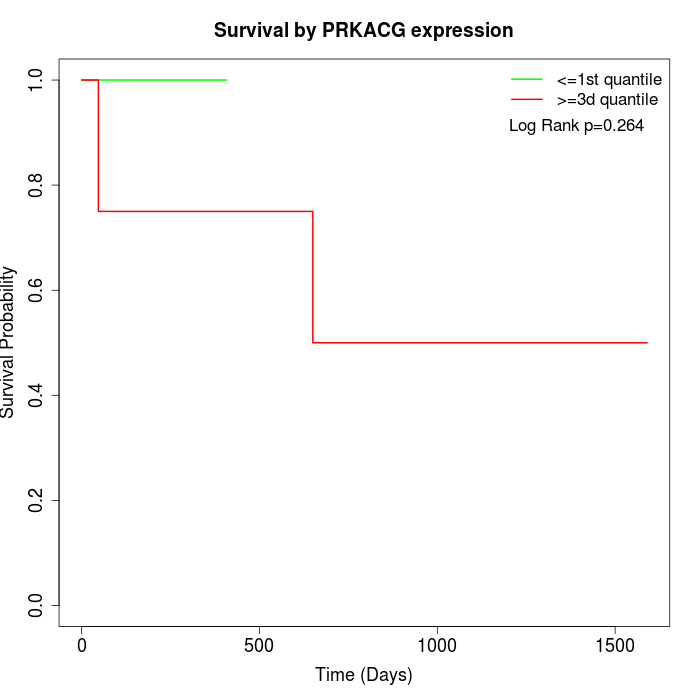
Survival by PRKACG expression:
Note: Click image to view full size file.
Copy number change of PRKACG:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PRKACG | 5568 | 6 | 10 | 14 | |
| GSE20123 | PRKACG | 5568 | 6 | 10 | 14 | |
| GSE43470 | PRKACG | 5568 | 6 | 5 | 32 | |
| GSE46452 | PRKACG | 5568 | 7 | 14 | 38 | |
| GSE47630 | PRKACG | 5568 | 1 | 19 | 20 | |
| GSE54993 | PRKACG | 5568 | 5 | 1 | 64 | |
| GSE54994 | PRKACG | 5568 | 4 | 13 | 36 | |
| GSE60625 | PRKACG | 5568 | 0 | 0 | 11 | |
| GSE74703 | PRKACG | 5568 | 5 | 4 | 27 | |
| GSE74704 | PRKACG | 5568 | 3 | 7 | 10 | |
| TCGA | PRKACG | 5568 | 18 | 28 | 50 |
Total number of gains: 61; Total number of losses: 111; Total Number of normals: 316.
Somatic mutations of PRKACG:
Generating mutation plots.
Highly correlated genes for PRKACG:
Showing top 20/716 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PRKACG | GYPC | 0.745506 | 3 | 0 | 3 |
| PRKACG | CNTF | 0.736745 | 3 | 0 | 3 |
| PRKACG | GRPR | 0.718424 | 4 | 0 | 4 |
| PRKACG | RUNX1T1 | 0.714544 | 3 | 0 | 3 |
| PRKACG | GABRA5 | 0.711846 | 3 | 0 | 3 |
| PRKACG | OR12D2 | 0.707489 | 4 | 0 | 4 |
| PRKACG | FSTL4 | 0.707431 | 4 | 0 | 4 |
| PRKACG | CGA | 0.706871 | 4 | 0 | 4 |
| PRKACG | GADL1 | 0.704147 | 3 | 0 | 3 |
| PRKACG | OR10J1 | 0.697492 | 3 | 0 | 3 |
| PRKACG | KAT7 | 0.696703 | 3 | 0 | 3 |
| PRKACG | ASH1L | 0.695475 | 3 | 0 | 3 |
| PRKACG | GDF2 | 0.693005 | 4 | 0 | 4 |
| PRKACG | MAP1A | 0.689224 | 4 | 0 | 3 |
| PRKACG | NTRK3 | 0.686091 | 4 | 0 | 4 |
| PRKACG | CCDC87 | 0.685181 | 4 | 0 | 4 |
| PRKACG | NTNG1 | 0.684969 | 3 | 0 | 3 |
| PRKACG | EVC2 | 0.684534 | 3 | 0 | 3 |
| PRKACG | SERPINA5 | 0.68319 | 5 | 0 | 5 |
| PRKACG | IQSEC2 | 0.682782 | 5 | 0 | 5 |
For details and further investigation, click here