| Full name: resistin like beta | Alias Symbol: HXCP2|FIZZ2|RELMb | ||
| Type: protein-coding gene | Cytoband: 3q13.13 | ||
| Entrez ID: 84666 | HGNC ID: HGNC:20388 | Ensembl Gene: ENSG00000163515 | OMIM ID: 605645 |
| Drug and gene relationship at DGIdb | |||
Expression of RETNLB:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | RETNLB | 84666 | 223969_s_at | 0.0291 | 0.9104 | |
| GSE26886 | RETNLB | 84666 | 223969_s_at | -0.0251 | 0.8113 | |
| GSE45670 | RETNLB | 84666 | 223969_s_at | 0.0880 | 0.3232 | |
| GSE53622 | RETNLB | 84666 | 74005 | 1.0110 | 0.0000 | |
| GSE53624 | RETNLB | 84666 | 74005 | 0.4922 | 0.0002 | |
| GSE63941 | RETNLB | 84666 | 223969_s_at | 0.0124 | 0.9520 | |
| GSE77861 | RETNLB | 84666 | 223969_s_at | -0.1055 | 0.1497 | |
| TCGA | RETNLB | 84666 | RNAseq | 0.6614 | 0.5938 |
Upregulated datasets: 1; Downregulated datasets: 0.
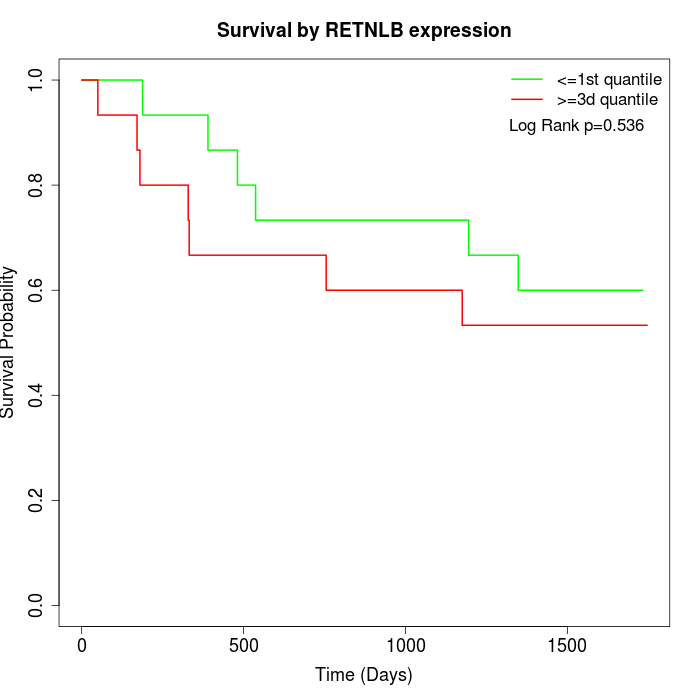
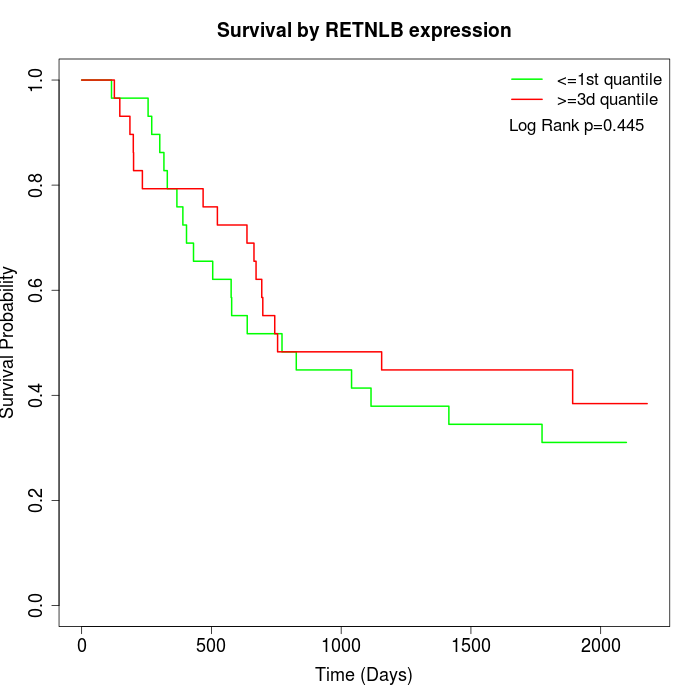
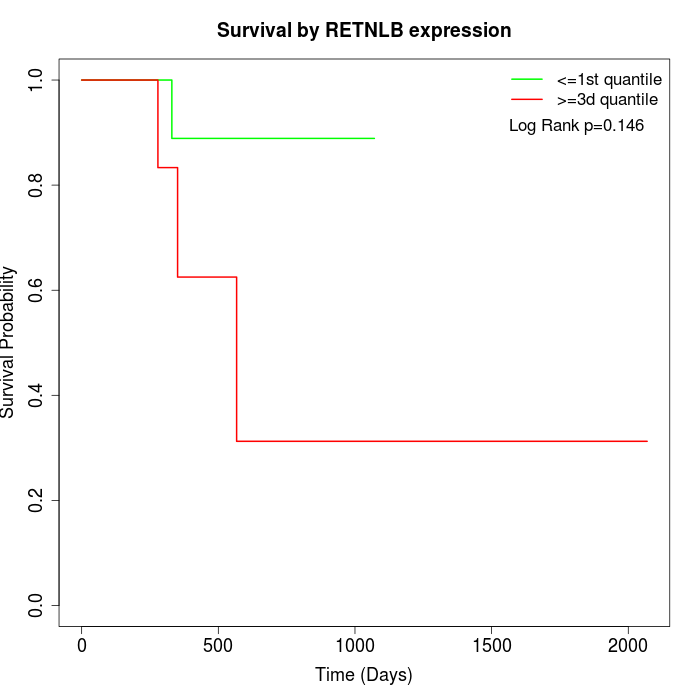
Survival by RETNLB expression:
Note: Click image to view full size file.
Copy number change of RETNLB:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RETNLB | 84666 | 17 | 1 | 12 | |
| GSE20123 | RETNLB | 84666 | 16 | 1 | 13 | |
| GSE43470 | RETNLB | 84666 | 18 | 1 | 24 | |
| GSE46452 | RETNLB | 84666 | 12 | 6 | 41 | |
| GSE47630 | RETNLB | 84666 | 16 | 6 | 18 | |
| GSE54993 | RETNLB | 84666 | 2 | 5 | 63 | |
| GSE54994 | RETNLB | 84666 | 28 | 5 | 20 | |
| GSE60625 | RETNLB | 84666 | 0 | 6 | 5 | |
| GSE74703 | RETNLB | 84666 | 15 | 1 | 20 | |
| GSE74704 | RETNLB | 84666 | 13 | 1 | 6 | |
| TCGA | RETNLB | 84666 | 50 | 8 | 38 |
Total number of gains: 187; Total number of losses: 41; Total Number of normals: 260.
Somatic mutations of RETNLB:
Generating mutation plots.
Highly correlated genes for RETNLB:
Showing top 20/42 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RETNLB | TNP2 | 0.678143 | 3 | 0 | 3 |
| RETNLB | FGF23 | 0.672515 | 3 | 0 | 3 |
| RETNLB | TIAF1 | 0.665382 | 3 | 0 | 3 |
| RETNLB | TMEM105 | 0.646184 | 3 | 0 | 3 |
| RETNLB | STAR | 0.637745 | 4 | 0 | 3 |
| RETNLB | FGF4 | 0.634783 | 3 | 0 | 3 |
| RETNLB | JARID2-AS1 | 0.629807 | 3 | 0 | 3 |
| RETNLB | DEFB118 | 0.621837 | 3 | 0 | 3 |
| RETNLB | NMUR2 | 0.616012 | 3 | 0 | 3 |
| RETNLB | HCRT | 0.607428 | 3 | 0 | 3 |
| RETNLB | RLBP1 | 0.601954 | 4 | 0 | 3 |
| RETNLB | MESP1 | 0.594313 | 4 | 0 | 3 |
| RETNLB | GLYAT | 0.58922 | 3 | 0 | 3 |
| RETNLB | CHRNA2 | 0.585891 | 5 | 0 | 4 |
| RETNLB | USH1G | 0.584098 | 3 | 0 | 3 |
| RETNLB | CCDC33 | 0.583334 | 5 | 0 | 4 |
| RETNLB | RGL4 | 0.572906 | 3 | 0 | 3 |
| RETNLB | ABI3 | 0.572053 | 3 | 0 | 3 |
| RETNLB | VGLL2 | 0.571245 | 3 | 0 | 3 |
| RETNLB | PLCH1-AS1 | 0.569415 | 4 | 0 | 3 |
For details and further investigation, click here