| Full name: glycine-N-acyltransferase | Alias Symbol: GAT|ACGNAT | ||
| Type: protein-coding gene | Cytoband: 11q12.1 | ||
| Entrez ID: 10249 | HGNC ID: HGNC:13734 | Ensembl Gene: ENSG00000149124 | OMIM ID: 607424 |
| Drug and gene relationship at DGIdb | |||
Expression of GLYAT:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GLYAT | 10249 | 222083_at | -0.3905 | 0.1374 | |
| GSE20347 | GLYAT | 10249 | 222083_at | -0.2160 | 0.0042 | |
| GSE23400 | GLYAT | 10249 | 222083_at | -0.3700 | 0.0000 | |
| GSE26886 | GLYAT | 10249 | 222083_at | -0.4798 | 0.0010 | |
| GSE29001 | GLYAT | 10249 | 222083_at | -0.3839 | 0.0275 | |
| GSE38129 | GLYAT | 10249 | 222083_at | -0.2716 | 0.0053 | |
| GSE45670 | GLYAT | 10249 | 222083_at | -0.1032 | 0.3123 | |
| GSE53622 | GLYAT | 10249 | 92447 | 0.5089 | 0.0016 | |
| GSE53624 | GLYAT | 10249 | 92447 | 0.4989 | 0.0000 | |
| GSE63941 | GLYAT | 10249 | 222083_at | -0.1668 | 0.3091 | |
| GSE77861 | GLYAT | 10249 | 222083_at | -0.3693 | 0.0161 | |
| SRP133303 | GLYAT | 10249 | RNAseq | 0.5953 | 0.0008 | |
| SRP159526 | GLYAT | 10249 | RNAseq | 0.8721 | 0.1686 | |
| SRP219564 | GLYAT | 10249 | RNAseq | 0.5684 | 0.0900 | |
| TCGA | GLYAT | 10249 | RNAseq | 0.6182 | 0.6866 |
Upregulated datasets: 0; Downregulated datasets: 0.
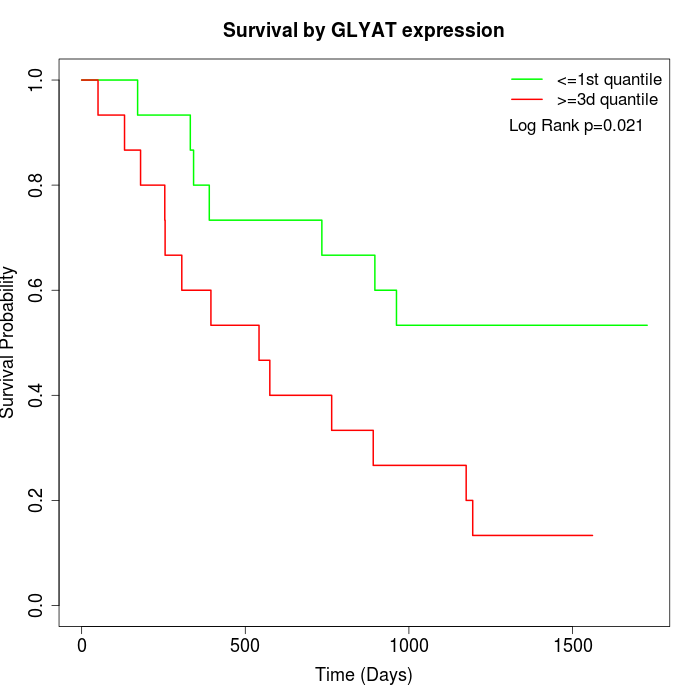
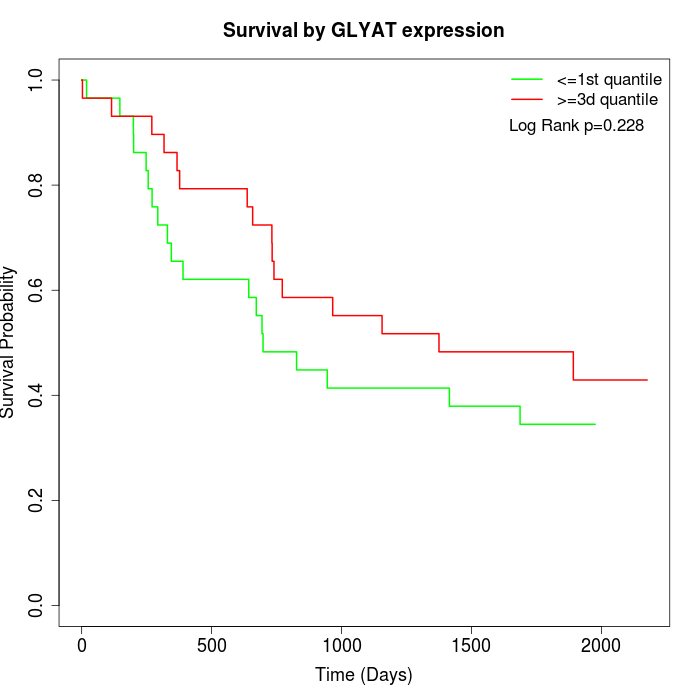
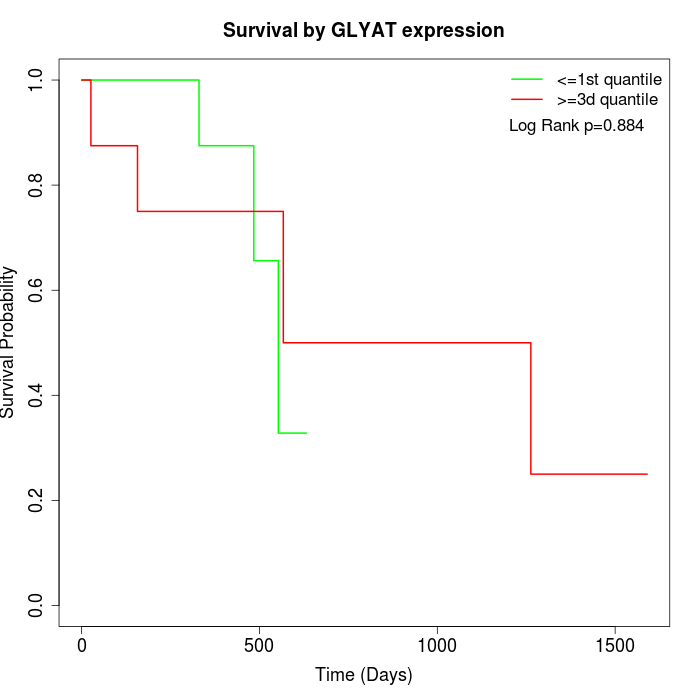
Survival by GLYAT expression:
Note: Click image to view full size file.
Copy number change of GLYAT:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GLYAT | 10249 | 1 | 4 | 25 | |
| GSE20123 | GLYAT | 10249 | 1 | 4 | 25 | |
| GSE43470 | GLYAT | 10249 | 1 | 5 | 37 | |
| GSE46452 | GLYAT | 10249 | 10 | 4 | 45 | |
| GSE47630 | GLYAT | 10249 | 3 | 8 | 29 | |
| GSE54993 | GLYAT | 10249 | 3 | 0 | 67 | |
| GSE54994 | GLYAT | 10249 | 5 | 6 | 42 | |
| GSE60625 | GLYAT | 10249 | 0 | 3 | 8 | |
| GSE74703 | GLYAT | 10249 | 1 | 3 | 32 | |
| GSE74704 | GLYAT | 10249 | 1 | 4 | 15 | |
| TCGA | GLYAT | 10249 | 14 | 10 | 72 |
Total number of gains: 40; Total number of losses: 51; Total Number of normals: 397.
Somatic mutations of GLYAT:
Generating mutation plots.
Highly correlated genes for GLYAT:
Showing top 20/1243 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GLYAT | FBXL16 | 0.762717 | 3 | 0 | 3 |
| GLYAT | NAV2-AS5 | 0.749389 | 3 | 0 | 3 |
| GLYAT | CYP19A1 | 0.740178 | 4 | 0 | 4 |
| GLYAT | RABEP2 | 0.732354 | 3 | 0 | 3 |
| GLYAT | PIK3R5 | 0.717567 | 4 | 0 | 4 |
| GLYAT | CAMK1G | 0.711099 | 5 | 0 | 5 |
| GLYAT | MVK | 0.698195 | 8 | 0 | 8 |
| GLYAT | SPX | 0.697082 | 6 | 0 | 6 |
| GLYAT | IL17A | 0.689234 | 4 | 0 | 4 |
| GLYAT | HHLA1 | 0.682014 | 7 | 0 | 7 |
| GLYAT | HOXA3 | 0.681626 | 5 | 0 | 5 |
| GLYAT | C2orf16 | 0.678712 | 4 | 0 | 4 |
| GLYAT | ZNF844 | 0.677723 | 3 | 0 | 3 |
| GLYAT | PSG2 | 0.677265 | 7 | 0 | 7 |
| GLYAT | ARSE | 0.675188 | 4 | 0 | 3 |
| GLYAT | BMP15 | 0.674093 | 5 | 0 | 5 |
| GLYAT | MAPK3 | 0.674033 | 8 | 0 | 7 |
| GLYAT | FAM13C | 0.673934 | 5 | 0 | 4 |
| GLYAT | THRB | 0.673861 | 4 | 0 | 4 |
| GLYAT | ESAM | 0.672911 | 3 | 0 | 3 |
For details and further investigation, click here