| Full name: fibroblast growth factor 4 | Alias Symbol: K-FGF|HBGF-4|HST|HST-1|KFGF | ||
| Type: protein-coding gene | Cytoband: 11q13.3 | ||
| Entrez ID: 2249 | HGNC ID: HGNC:3682 | Ensembl Gene: ENSG00000075388 | OMIM ID: 164980 |
| Drug and gene relationship at DGIdb | |||
FGF4 involved pathways:
Expression of FGF4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | FGF4 | 2249 | 206783_at | -0.1576 | 0.4859 | |
| GSE20347 | FGF4 | 2249 | 206783_at | 0.1146 | 0.3340 | |
| GSE23400 | FGF4 | 2249 | 206783_at | -0.0618 | 0.1440 | |
| GSE26886 | FGF4 | 2249 | 1552982_a_at | 0.1059 | 0.3551 | |
| GSE29001 | FGF4 | 2249 | 206783_at | -0.1731 | 0.3071 | |
| GSE38129 | FGF4 | 2249 | 206783_at | -0.0476 | 0.7987 | |
| GSE45670 | FGF4 | 2249 | 206783_at | 0.1110 | 0.2584 | |
| GSE53622 | FGF4 | 2249 | 22440 | 0.5024 | 0.0000 | |
| GSE53624 | FGF4 | 2249 | 22440 | 0.6385 | 0.0000 | |
| GSE63941 | FGF4 | 2249 | 206783_at | 0.1000 | 0.5385 | |
| GSE77861 | FGF4 | 2249 | 206783_at | -0.0965 | 0.3569 | |
| TCGA | FGF4 | 2249 | RNAseq | 2.1656 | 0.2931 |
Upregulated datasets: 0; Downregulated datasets: 0.
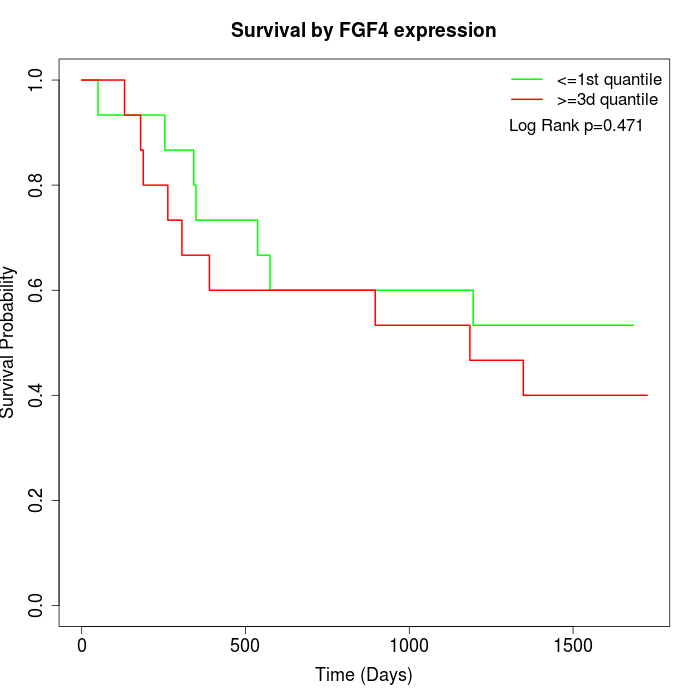
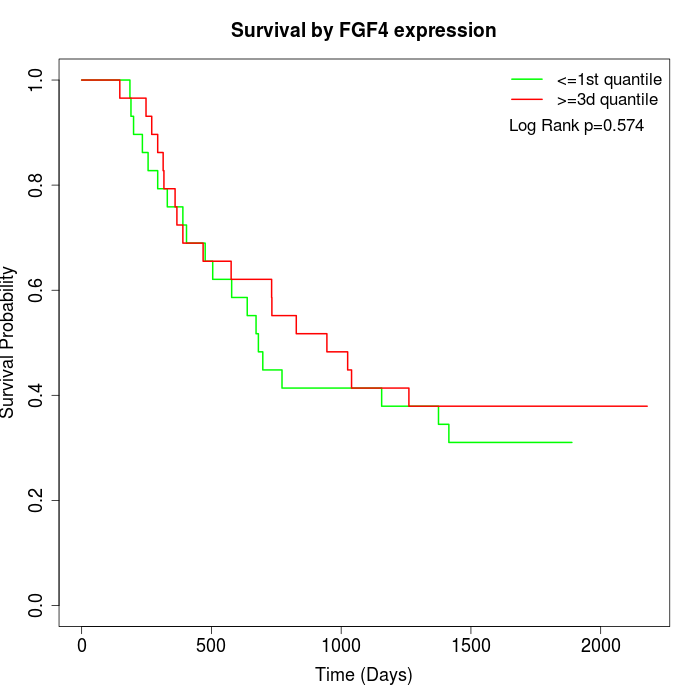
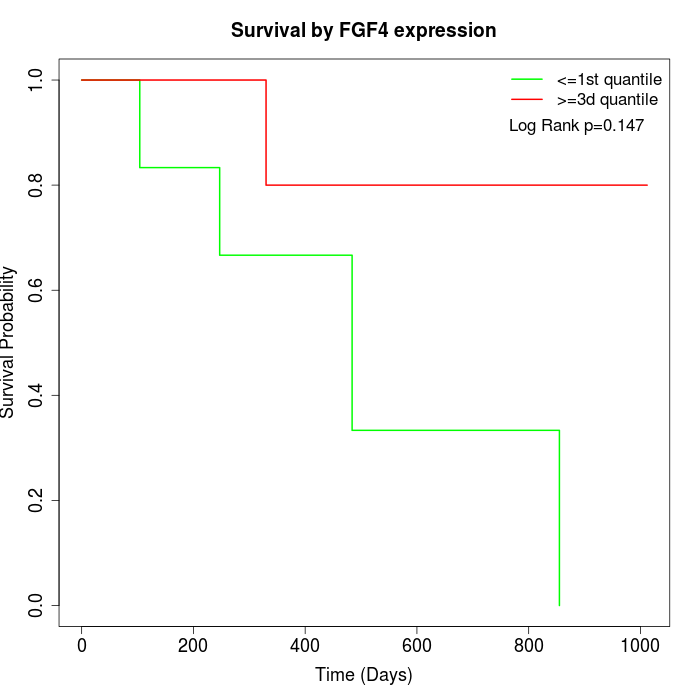
Survival by FGF4 expression:
Note: Click image to view full size file.
Copy number change of FGF4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | FGF4 | 2249 | 19 | 1 | 10 | |
| GSE20123 | FGF4 | 2249 | 19 | 1 | 10 | |
| GSE43470 | FGF4 | 2249 | 16 | 2 | 25 | |
| GSE46452 | FGF4 | 2249 | 36 | 1 | 22 | |
| GSE47630 | FGF4 | 2249 | 20 | 1 | 19 | |
| GSE54993 | FGF4 | 2249 | 1 | 25 | 44 | |
| GSE54994 | FGF4 | 2249 | 28 | 3 | 22 | |
| GSE60625 | FGF4 | 2249 | 0 | 8 | 3 | |
| GSE74703 | FGF4 | 2249 | 13 | 1 | 22 | |
| GSE74704 | FGF4 | 2249 | 13 | 0 | 7 | |
| TCGA | FGF4 | 2249 | 63 | 2 | 31 |
Total number of gains: 228; Total number of losses: 45; Total Number of normals: 215.
Somatic mutations of FGF4:
Generating mutation plots.
Highly correlated genes for FGF4:
Showing top 20/552 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| FGF4 | TBXA2R | 0.73745 | 4 | 0 | 4 |
| FGF4 | TULP2 | 0.726593 | 3 | 0 | 3 |
| FGF4 | MUC13 | 0.721192 | 3 | 0 | 3 |
| FGF4 | TRPM8 | 0.717999 | 3 | 0 | 3 |
| FGF4 | AGTR2 | 0.700384 | 4 | 0 | 3 |
| FGF4 | KCNMB2 | 0.691407 | 5 | 0 | 4 |
| FGF4 | NOX5 | 0.688528 | 3 | 0 | 3 |
| FGF4 | STXBP6 | 0.686358 | 3 | 0 | 3 |
| FGF4 | PIWIL2 | 0.685841 | 4 | 0 | 4 |
| FGF4 | LIMD1-AS1 | 0.682379 | 3 | 0 | 3 |
| FGF4 | MLNR | 0.675814 | 3 | 0 | 3 |
| FGF4 | NR0B2 | 0.673251 | 4 | 0 | 4 |
| FGF4 | GABRA1 | 0.672205 | 4 | 0 | 4 |
| FGF4 | OPRK1 | 0.66939 | 3 | 0 | 3 |
| FGF4 | KRTAP4-7 | 0.667932 | 3 | 0 | 3 |
| FGF4 | VWA7 | 0.667302 | 5 | 0 | 4 |
| FGF4 | RHAG | 0.665056 | 3 | 0 | 3 |
| FGF4 | FASLG | 0.662853 | 3 | 0 | 3 |
| FGF4 | ATP8A2 | 0.661128 | 5 | 0 | 5 |
| FGF4 | ARHGAP44 | 0.658971 | 3 | 0 | 3 |
For details and further investigation, click here