| Full name: neuromedin U receptor 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 5q33.1 | ||
| Entrez ID: 56923 | HGNC ID: HGNC:16454 | Ensembl Gene: ENSG00000132911 | OMIM ID: 605108 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of NMUR2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NMUR2 | 56923 | 224088_at | 0.0262 | 0.9479 | |
| GSE26886 | NMUR2 | 56923 | 224088_at | -0.2398 | 0.1001 | |
| GSE45670 | NMUR2 | 56923 | 224088_at | 0.1639 | 0.1884 | |
| GSE53622 | NMUR2 | 56923 | 13217 | 0.5206 | 0.0002 | |
| GSE53624 | NMUR2 | 56923 | 13217 | 0.7120 | 0.0000 | |
| GSE63941 | NMUR2 | 56923 | 224088_at | 0.0822 | 0.6874 | |
| GSE77861 | NMUR2 | 56923 | 224088_at | -0.1625 | 0.1256 | |
| GSE97050 | NMUR2 | 56923 | A_23_P122134 | -0.1083 | 0.6706 | |
| TCGA | NMUR2 | 56923 | RNAseq | 0.0442 | 0.9524 |
Upregulated datasets: 0; Downregulated datasets: 0.
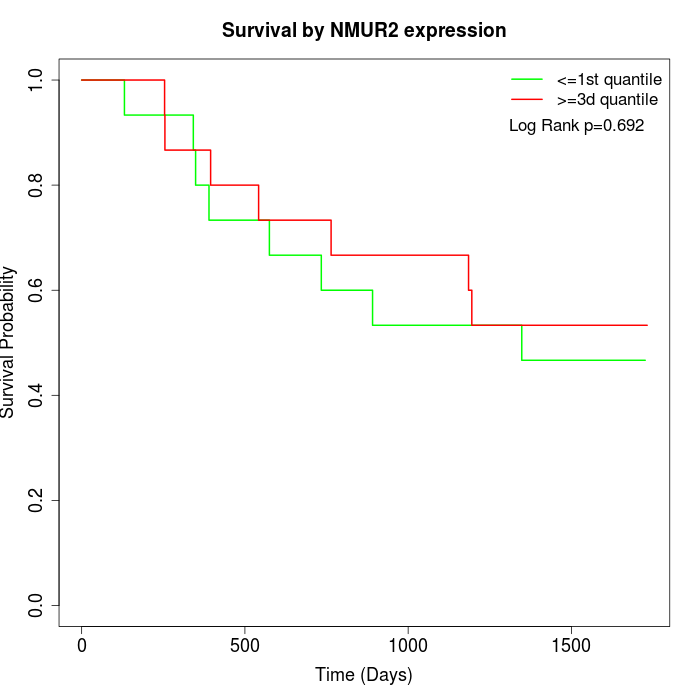
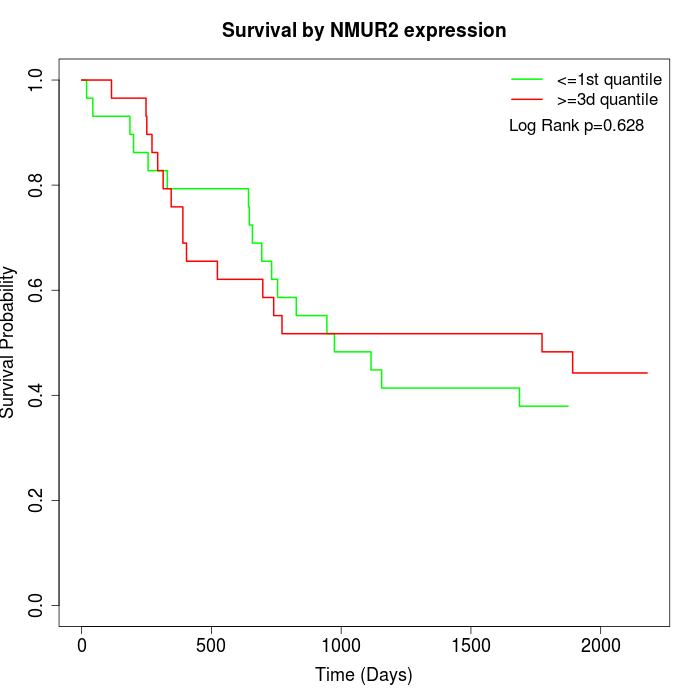
Survival by NMUR2 expression:
Note: Click image to view full size file.
Copy number change of NMUR2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NMUR2 | 56923 | 1 | 12 | 17 | |
| GSE20123 | NMUR2 | 56923 | 1 | 12 | 17 | |
| GSE43470 | NMUR2 | 56923 | 2 | 10 | 31 | |
| GSE46452 | NMUR2 | 56923 | 0 | 27 | 32 | |
| GSE47630 | NMUR2 | 56923 | 0 | 20 | 20 | |
| GSE54993 | NMUR2 | 56923 | 9 | 1 | 60 | |
| GSE54994 | NMUR2 | 56923 | 1 | 16 | 36 | |
| GSE60625 | NMUR2 | 56923 | 0 | 0 | 11 | |
| GSE74703 | NMUR2 | 56923 | 2 | 8 | 26 | |
| GSE74704 | NMUR2 | 56923 | 1 | 7 | 12 | |
| TCGA | NMUR2 | 56923 | 5 | 37 | 54 |
Total number of gains: 22; Total number of losses: 150; Total Number of normals: 316.
Somatic mutations of NMUR2:
Generating mutation plots.
Highly correlated genes for NMUR2:
Showing top 20/136 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NMUR2 | HAND1 | 0.724101 | 3 | 0 | 3 |
| NMUR2 | TTR | 0.709702 | 3 | 0 | 3 |
| NMUR2 | AQP9 | 0.675949 | 3 | 0 | 3 |
| NMUR2 | KIRREL3-AS3 | 0.667431 | 4 | 0 | 4 |
| NMUR2 | CELA3A | 0.664458 | 4 | 0 | 4 |
| NMUR2 | EPOR | 0.661829 | 4 | 0 | 3 |
| NMUR2 | KLLN | 0.660773 | 4 | 0 | 3 |
| NMUR2 | GLT1D1 | 0.656666 | 3 | 0 | 3 |
| NMUR2 | PRSS33 | 0.646403 | 3 | 0 | 3 |
| NMUR2 | CSRP3 | 0.637163 | 3 | 0 | 3 |
| NMUR2 | LINC01192 | 0.635996 | 3 | 0 | 3 |
| NMUR2 | ANHX | 0.63301 | 4 | 0 | 3 |
| NMUR2 | KRT82 | 0.631454 | 3 | 0 | 3 |
| NMUR2 | GPR75 | 0.628569 | 3 | 0 | 3 |
| NMUR2 | FLVCR2 | 0.628351 | 3 | 0 | 3 |
| NMUR2 | MPV17L | 0.624929 | 4 | 0 | 4 |
| NMUR2 | SPATA32 | 0.624436 | 3 | 0 | 3 |
| NMUR2 | ASCL3 | 0.619868 | 4 | 0 | 3 |
| NMUR2 | RETNLB | 0.616012 | 3 | 0 | 3 |
| NMUR2 | DCST1 | 0.614185 | 3 | 0 | 3 |
For details and further investigation, click here