| Full name: hypocretin neuropeptide precursor | Alias Symbol: PPOX|OX | ||
| Type: protein-coding gene | Cytoband: 17q21.2 | ||
| Entrez ID: 3060 | HGNC ID: HGNC:4847 | Ensembl Gene: ENSG00000161610 | OMIM ID: 602358 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of HCRT:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | HCRT | 3060 | 207642_at | -0.3732 | 0.1369 | |
| GSE20347 | HCRT | 3060 | 207642_at | -0.1623 | 0.0164 | |
| GSE23400 | HCRT | 3060 | 207642_at | -0.2353 | 0.0000 | |
| GSE26886 | HCRT | 3060 | 207642_at | 0.0472 | 0.8123 | |
| GSE29001 | HCRT | 3060 | 207642_at | -0.2707 | 0.0426 | |
| GSE38129 | HCRT | 3060 | 207642_at | -0.2807 | 0.0000 | |
| GSE45670 | HCRT | 3060 | 207642_at | -0.1039 | 0.1981 | |
| GSE53622 | HCRT | 3060 | 84358 | -0.2893 | 0.0044 | |
| GSE53624 | HCRT | 3060 | 84358 | -0.4779 | 0.0002 | |
| GSE63941 | HCRT | 3060 | 207642_at | 0.1575 | 0.3208 | |
| GSE77861 | HCRT | 3060 | 207642_at | -0.2487 | 0.1604 | |
| GSE97050 | HCRT | 3060 | A_33_P3306624 | 0.0969 | 0.6212 |
Upregulated datasets: 0; Downregulated datasets: 0.
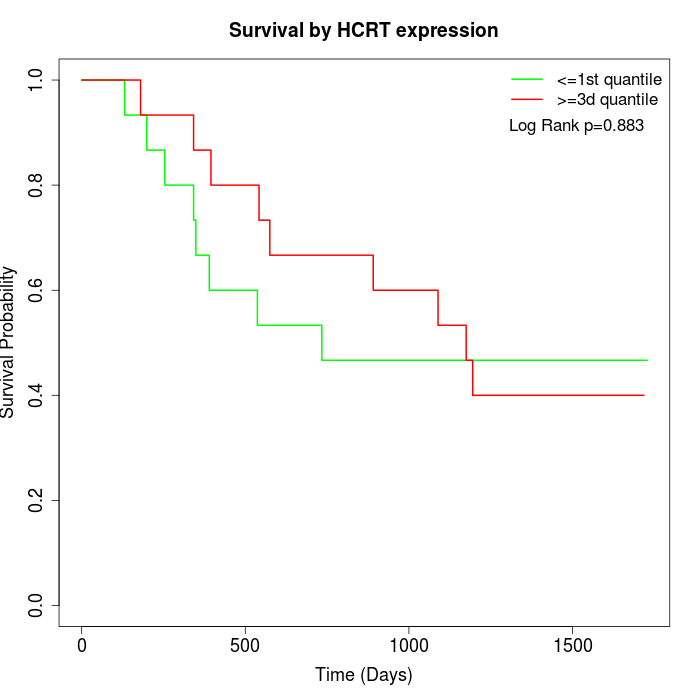
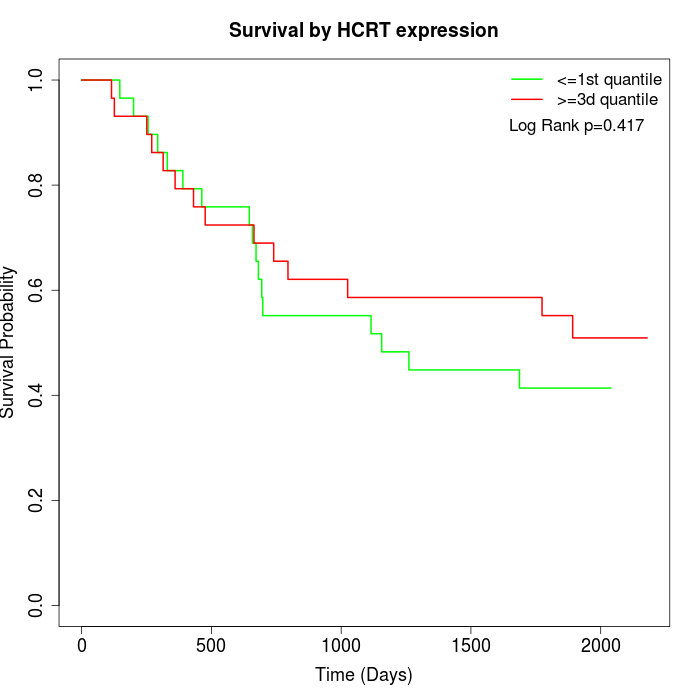
Survival by HCRT expression:
Note: Click image to view full size file.
Copy number change of HCRT:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | HCRT | 3060 | 6 | 2 | 22 | |
| GSE20123 | HCRT | 3060 | 6 | 2 | 22 | |
| GSE43470 | HCRT | 3060 | 1 | 2 | 40 | |
| GSE46452 | HCRT | 3060 | 34 | 0 | 25 | |
| GSE47630 | HCRT | 3060 | 8 | 1 | 31 | |
| GSE54993 | HCRT | 3060 | 3 | 4 | 63 | |
| GSE54994 | HCRT | 3060 | 8 | 5 | 40 | |
| GSE60625 | HCRT | 3060 | 4 | 0 | 7 | |
| GSE74703 | HCRT | 3060 | 1 | 1 | 34 | |
| GSE74704 | HCRT | 3060 | 4 | 1 | 15 | |
| TCGA | HCRT | 3060 | 23 | 6 | 67 |
Total number of gains: 98; Total number of losses: 24; Total Number of normals: 366.
Somatic mutations of HCRT:
Generating mutation plots.
Highly correlated genes for HCRT:
Showing top 20/1062 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| HCRT | SLC25A48 | 0.81098 | 3 | 0 | 3 |
| HCRT | CELF5 | 0.789539 | 3 | 0 | 3 |
| HCRT | ZNF491 | 0.787576 | 3 | 0 | 3 |
| HCRT | MRGPRE | 0.775079 | 3 | 0 | 3 |
| HCRT | PRAMEF12 | 0.766308 | 3 | 0 | 3 |
| HCRT | CALCA | 0.765108 | 5 | 0 | 5 |
| HCRT | NXNL1 | 0.764755 | 3 | 0 | 3 |
| HCRT | TSPAN10 | 0.757607 | 3 | 0 | 3 |
| HCRT | INO80C | 0.756191 | 3 | 0 | 3 |
| HCRT | KRTAP6-1 | 0.754119 | 3 | 0 | 3 |
| HCRT | POU4F3 | 0.754094 | 4 | 0 | 4 |
| HCRT | CRB2 | 0.750012 | 3 | 0 | 3 |
| HCRT | NLRP5 | 0.736993 | 3 | 0 | 3 |
| HCRT | CNPY1 | 0.735638 | 4 | 0 | 3 |
| HCRT | ANKRD33B | 0.73466 | 3 | 0 | 3 |
| HCRT | C2orf50 | 0.728273 | 3 | 0 | 3 |
| HCRT | VSTM2A | 0.718529 | 4 | 0 | 4 |
| HCRT | INHBC | 0.716267 | 3 | 0 | 3 |
| HCRT | PPP1R14D | 0.715866 | 5 | 0 | 5 |
| HCRT | KCNMB2 | 0.709441 | 5 | 0 | 5 |
For details and further investigation, click here