| Full name: solute carrier family 4 member 9 | Alias Symbol: AE4 | ||
| Type: protein-coding gene | Cytoband: 5q31.3 | ||
| Entrez ID: 83697 | HGNC ID: HGNC:11035 | Ensembl Gene: ENSG00000113073 | OMIM ID: 610207 |
| Drug and gene relationship at DGIdb | |||
Expression of SLC4A9:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SLC4A9 | 83697 | 224053_s_at | -0.1318 | 0.5614 | |
| GSE26886 | SLC4A9 | 83697 | 224053_s_at | 0.0418 | 0.7355 | |
| GSE45670 | SLC4A9 | 83697 | 224053_s_at | 0.0002 | 0.9988 | |
| GSE53622 | SLC4A9 | 83697 | 29439 | -0.5228 | 0.0023 | |
| GSE53624 | SLC4A9 | 83697 | 29439 | -1.2729 | 0.0000 | |
| GSE63941 | SLC4A9 | 83697 | 224053_s_at | 0.1324 | 0.3139 | |
| GSE77861 | SLC4A9 | 83697 | 224053_s_at | -0.0805 | 0.5326 | |
| GSE97050 | SLC4A9 | 83697 | A_23_P257176 | 0.1619 | 0.4762 | |
| TCGA | SLC4A9 | 83697 | RNAseq | 2.6051 | 0.0007 |
Upregulated datasets: 1; Downregulated datasets: 1.
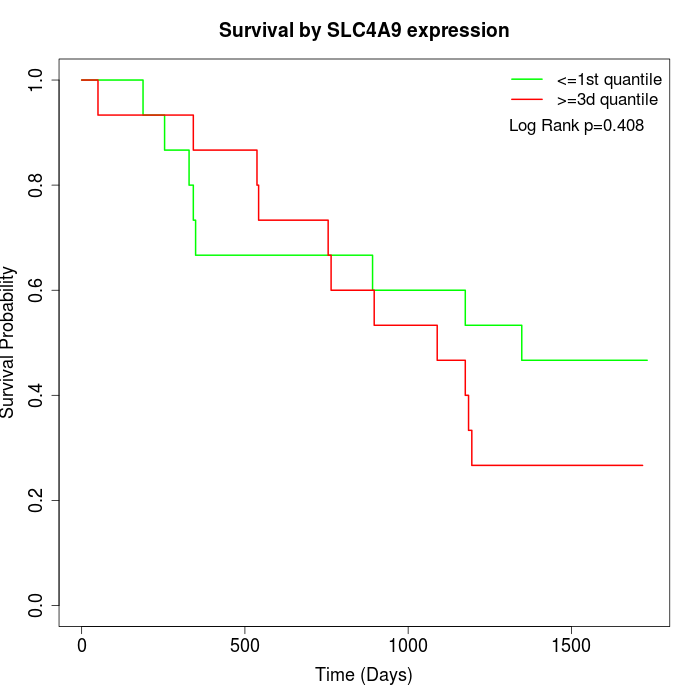
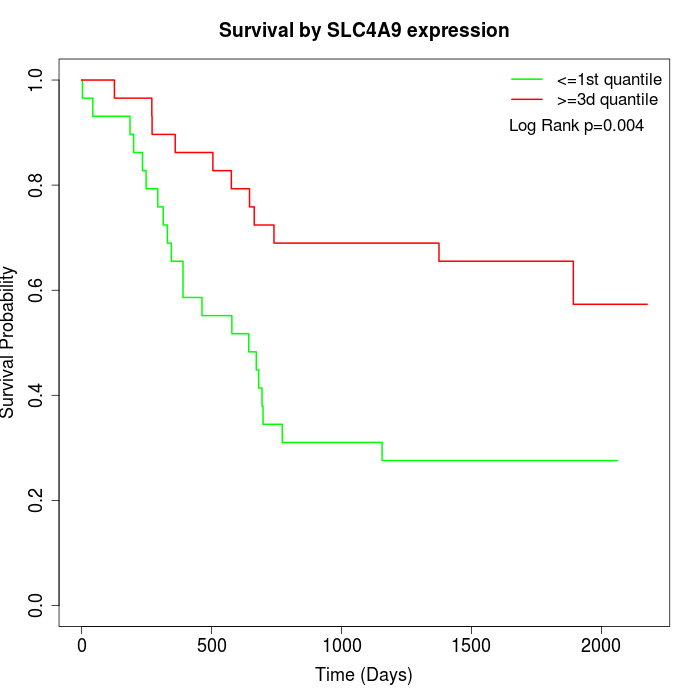
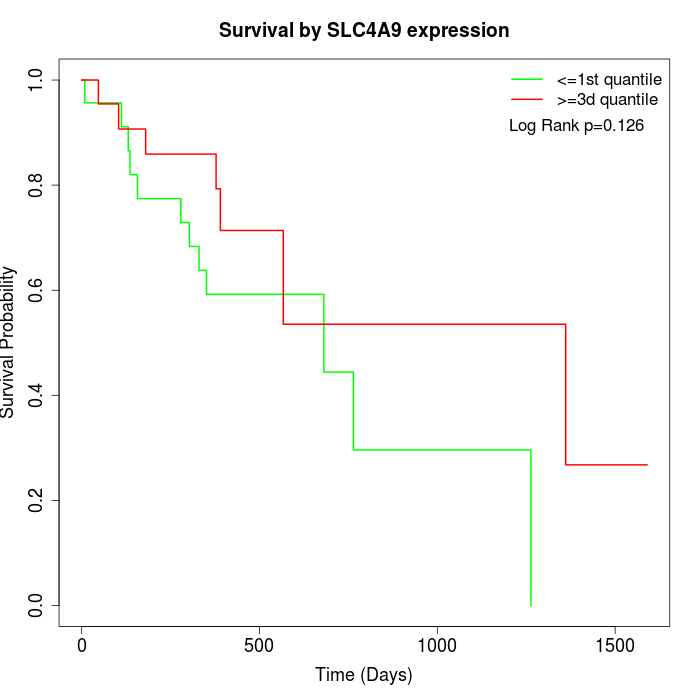
Survival by SLC4A9 expression:
Note: Click image to view full size file.
Copy number change of SLC4A9:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SLC4A9 | 83697 | 3 | 11 | 16 | |
| GSE20123 | SLC4A9 | 83697 | 4 | 11 | 15 | |
| GSE43470 | SLC4A9 | 83697 | 2 | 10 | 31 | |
| GSE46452 | SLC4A9 | 83697 | 0 | 27 | 32 | |
| GSE47630 | SLC4A9 | 83697 | 0 | 20 | 20 | |
| GSE54993 | SLC4A9 | 83697 | 9 | 1 | 60 | |
| GSE54994 | SLC4A9 | 83697 | 2 | 14 | 37 | |
| GSE60625 | SLC4A9 | 83697 | 0 | 0 | 11 | |
| GSE74703 | SLC4A9 | 83697 | 2 | 7 | 27 | |
| GSE74704 | SLC4A9 | 83697 | 3 | 5 | 12 | |
| TCGA | SLC4A9 | 83697 | 4 | 37 | 55 |
Total number of gains: 29; Total number of losses: 143; Total Number of normals: 316.
Somatic mutations of SLC4A9:
Generating mutation plots.
Highly correlated genes for SLC4A9:
Showing top 20/491 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SLC4A9 | ZFR2 | 0.768641 | 3 | 0 | 3 |
| SLC4A9 | SPRR2F | 0.76387 | 3 | 0 | 3 |
| SLC4A9 | SPRR2D | 0.759616 | 3 | 0 | 3 |
| SLC4A9 | SPRR2A | 0.758544 | 3 | 0 | 3 |
| SLC4A9 | SPRR2E | 0.758174 | 3 | 0 | 3 |
| SLC4A9 | NKAIN4 | 0.750409 | 3 | 0 | 3 |
| SLC4A9 | FGG | 0.748721 | 3 | 0 | 3 |
| SLC4A9 | NCR1 | 0.729747 | 3 | 0 | 3 |
| SLC4A9 | RNF222 | 0.724707 | 3 | 0 | 3 |
| SLC4A9 | SLC25A48 | 0.716946 | 3 | 0 | 3 |
| SLC4A9 | MFSD2B | 0.712011 | 3 | 0 | 3 |
| SLC4A9 | KRTAP20-1 | 0.711594 | 3 | 0 | 3 |
| SLC4A9 | ISLR2 | 0.710393 | 3 | 0 | 3 |
| SLC4A9 | AMN | 0.709148 | 4 | 0 | 4 |
| SLC4A9 | BARHL2 | 0.707137 | 3 | 0 | 3 |
| SLC4A9 | CD164L2 | 0.706179 | 3 | 0 | 3 |
| SLC4A9 | FAM163B | 0.705776 | 3 | 0 | 3 |
| SLC4A9 | ARHGAP30 | 0.70326 | 3 | 0 | 3 |
| SLC4A9 | THEG | 0.703117 | 3 | 0 | 3 |
| SLC4A9 | OTOP3 | 0.699019 | 3 | 0 | 3 |
For details and further investigation, click here