| Full name: shadow of prion protein | Alias Symbol: Shadoo|Sprn|bA108K14.1|FLJ41197 | ||
| Type: protein-coding gene | Cytoband: 10q26.3 | ||
| Entrez ID: 503542 | HGNC ID: HGNC:16871 | Ensembl Gene: ENSG00000203772 | OMIM ID: 610447 |
| Drug and gene relationship at DGIdb | |||
Expression of SPRN:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SPRN | 503542 | 238331_at | 0.0445 | 0.8580 | |
| GSE26886 | SPRN | 503542 | 238331_at | 0.0510 | 0.7699 | |
| GSE45670 | SPRN | 503542 | 238331_at | 0.2088 | 0.0118 | |
| GSE53622 | SPRN | 503542 | 92963 | 0.3580 | 0.0000 | |
| GSE53624 | SPRN | 503542 | 92963 | 0.2900 | 0.0002 | |
| GSE63941 | SPRN | 503542 | 238331_at | 0.0339 | 0.8434 | |
| GSE77861 | SPRN | 503542 | 238331_at | -0.0799 | 0.5196 | |
| GSE97050 | SPRN | 503542 | A_24_P576506 | 0.0458 | 0.8759 | |
| SRP133303 | SPRN | 503542 | RNAseq | 0.1491 | 0.5428 | |
| SRP159526 | SPRN | 503542 | RNAseq | 2.4397 | 0.0001 | |
| TCGA | SPRN | 503542 | RNAseq | -0.3565 | 0.0044 |
Upregulated datasets: 1; Downregulated datasets: 0.
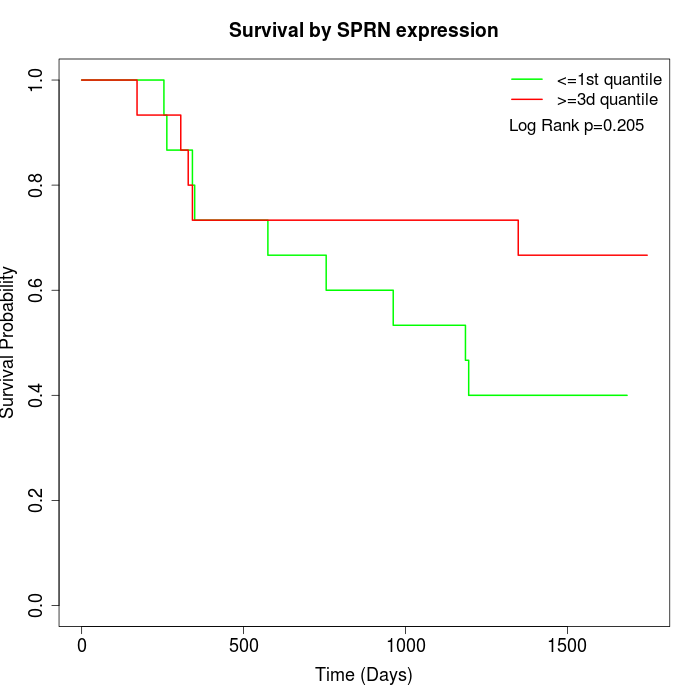
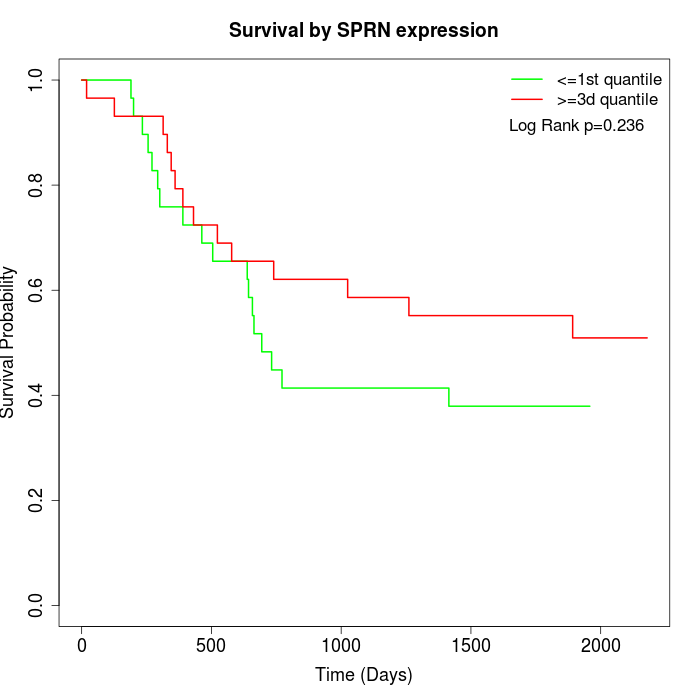
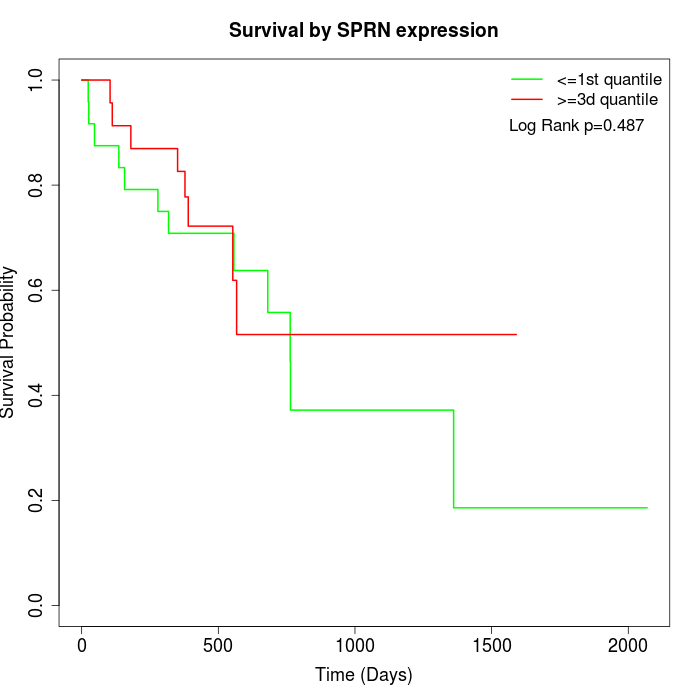
Survival by SPRN expression:
Note: Click image to view full size file.
Copy number change of SPRN:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SPRN | 503542 | 3 | 8 | 19 | |
| GSE20123 | SPRN | 503542 | 3 | 8 | 19 | |
| GSE43470 | SPRN | 503542 | 0 | 9 | 34 | |
| GSE46452 | SPRN | 503542 | 1 | 11 | 47 | |
| GSE47630 | SPRN | 503542 | 4 | 13 | 23 | |
| GSE54993 | SPRN | 503542 | 8 | 2 | 60 | |
| GSE54994 | SPRN | 503542 | 1 | 9 | 43 | |
| GSE60625 | SPRN | 503542 | 0 | 0 | 11 | |
| GSE74703 | SPRN | 503542 | 0 | 7 | 29 | |
| GSE74704 | SPRN | 503542 | 3 | 4 | 13 | |
| TCGA | SPRN | 503542 | 7 | 28 | 61 |
Total number of gains: 30; Total number of losses: 99; Total Number of normals: 359.
Somatic mutations of SPRN:
Generating mutation plots.
Highly correlated genes for SPRN:
Showing top 20/192 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SPRN | NTNG2 | 0.758591 | 3 | 0 | 3 |
| SPRN | CHST10 | 0.744428 | 3 | 0 | 3 |
| SPRN | PHF21A | 0.743713 | 3 | 0 | 3 |
| SPRN | CDH4 | 0.739326 | 3 | 0 | 3 |
| SPRN | SBNO2 | 0.732303 | 3 | 0 | 3 |
| SPRN | TMEM179 | 0.722784 | 3 | 0 | 3 |
| SPRN | PAX3 | 0.704905 | 3 | 0 | 3 |
| SPRN | GRIN2C | 0.698475 | 3 | 0 | 3 |
| SPRN | TMEM105 | 0.693884 | 4 | 0 | 4 |
| SPRN | EPB42 | 0.693175 | 3 | 0 | 3 |
| SPRN | PRLH | 0.690095 | 3 | 0 | 3 |
| SPRN | LENG9 | 0.687652 | 3 | 0 | 3 |
| SPRN | TBKBP1 | 0.686331 | 3 | 0 | 3 |
| SPRN | ZNF646 | 0.684678 | 3 | 0 | 3 |
| SPRN | CDC42EP2 | 0.678922 | 3 | 0 | 3 |
| SPRN | KIRREL2 | 0.677468 | 4 | 0 | 3 |
| SPRN | NXNL1 | 0.675472 | 4 | 0 | 4 |
| SPRN | NPR2 | 0.675011 | 3 | 0 | 3 |
| SPRN | RASL10A | 0.67412 | 5 | 0 | 5 |
| SPRN | MEPE | 0.673641 | 4 | 0 | 3 |
For details and further investigation, click here