| Full name: src-related kinase lacking C-terminal regulatory tyrosine and N-terminal myristylation sites | Alias Symbol: SRM|dJ697K14.1|PTK70 | ||
| Type: protein-coding gene | Cytoband: 20q13.33 | ||
| Entrez ID: 6725 | HGNC ID: HGNC:11298 | Ensembl Gene: ENSG00000125508 | OMIM ID: 617797 |
| Drug and gene relationship at DGIdb | |||
Expression of SRMS:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SRMS | 6725 | 234514_at | -0.2033 | 0.4001 | |
| GSE26886 | SRMS | 6725 | 234514_at | 0.1939 | 0.1625 | |
| GSE45670 | SRMS | 6725 | 234514_at | -0.1416 | 0.0874 | |
| GSE53622 | SRMS | 6725 | 7189 | 0.0622 | 0.5651 | |
| GSE53624 | SRMS | 6725 | 7189 | -0.1257 | 0.1640 | |
| GSE63941 | SRMS | 6725 | 234514_at | 0.1547 | 0.3091 | |
| GSE77861 | SRMS | 6725 | 234514_at | -0.0664 | 0.6215 | |
| GSE97050 | SRMS | 6725 | A_33_P3236798 | 0.3238 | 0.2740 | |
| SRP133303 | SRMS | 6725 | RNAseq | -0.9387 | 0.0329 | |
| SRP159526 | SRMS | 6725 | RNAseq | 0.6045 | 0.5578 | |
| TCGA | SRMS | 6725 | RNAseq | -1.0312 | 0.1462 |
Upregulated datasets: 0; Downregulated datasets: 0.
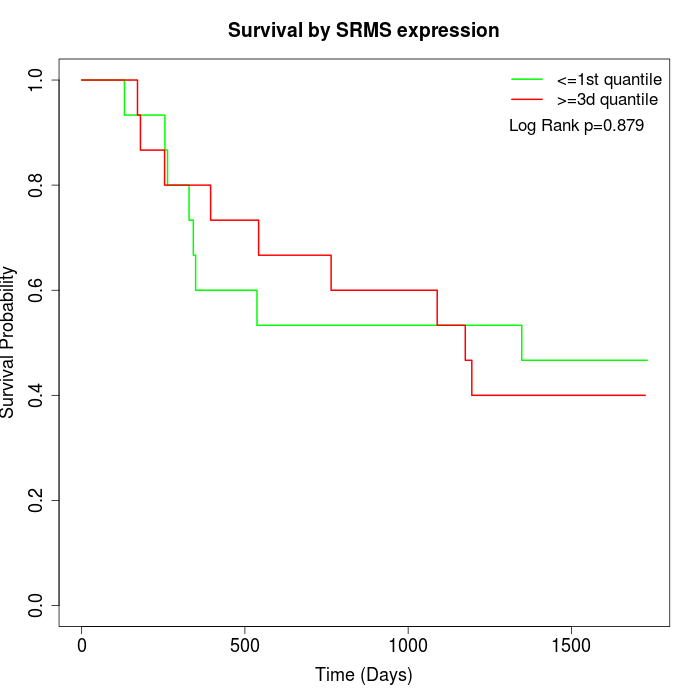
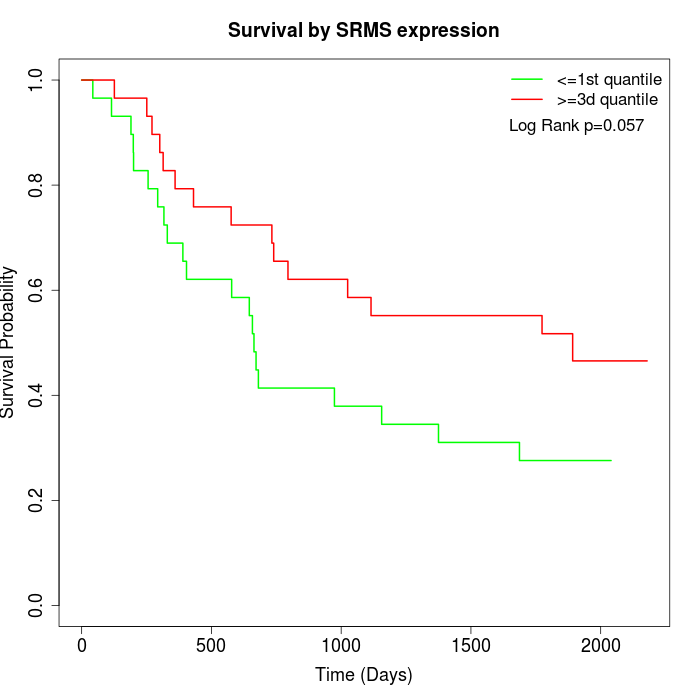
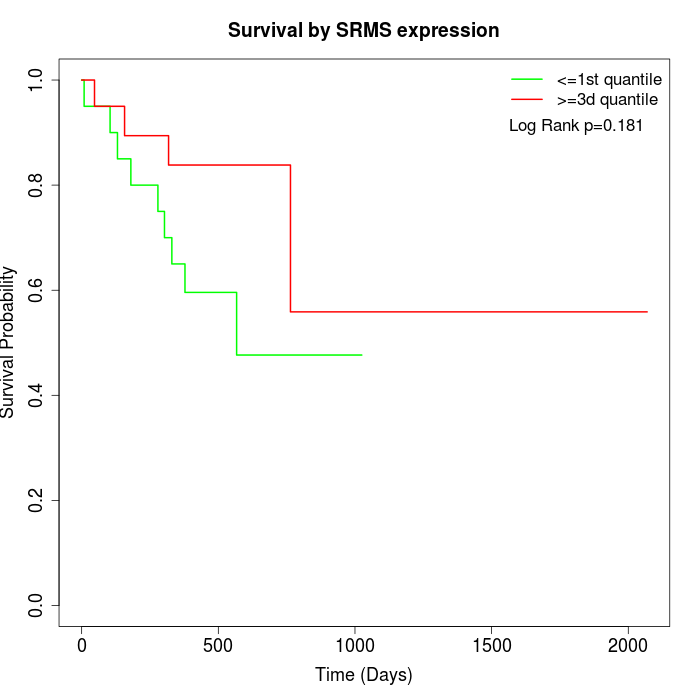
Survival by SRMS expression:
Note: Click image to view full size file.
Copy number change of SRMS:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SRMS | 6725 | 14 | 2 | 14 | |
| GSE20123 | SRMS | 6725 | 14 | 2 | 14 | |
| GSE43470 | SRMS | 6725 | 11 | 1 | 31 | |
| GSE46452 | SRMS | 6725 | 35 | 0 | 24 | |
| GSE47630 | SRMS | 6725 | 25 | 1 | 14 | |
| GSE54993 | SRMS | 6725 | 0 | 17 | 53 | |
| GSE54994 | SRMS | 6725 | 29 | 0 | 24 | |
| GSE60625 | SRMS | 6725 | 0 | 0 | 11 | |
| GSE74703 | SRMS | 6725 | 10 | 0 | 26 | |
| GSE74704 | SRMS | 6725 | 11 | 0 | 9 | |
| TCGA | SRMS | 6725 | 44 | 4 | 48 |
Total number of gains: 193; Total number of losses: 27; Total Number of normals: 268.
Somatic mutations of SRMS:
Generating mutation plots.
Highly correlated genes for SRMS:
Showing top 20/56 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SRMS | POU2F2 | 0.764107 | 3 | 0 | 3 |
| SRMS | PSKH1 | 0.704205 | 3 | 0 | 3 |
| SRMS | LINC00529 | 0.679119 | 3 | 0 | 3 |
| SRMS | CIITA | 0.670146 | 3 | 0 | 3 |
| SRMS | FUT5 | 0.657219 | 3 | 0 | 3 |
| SRMS | NMUR1 | 0.640881 | 3 | 0 | 3 |
| SRMS | CCR10 | 0.640061 | 3 | 0 | 3 |
| SRMS | RBPJL | 0.632591 | 3 | 0 | 3 |
| SRMS | RNF31 | 0.611757 | 4 | 0 | 3 |
| SRMS | C1orf127 | 0.605713 | 4 | 0 | 3 |
| SRMS | TAS2R45 | 0.605619 | 4 | 0 | 3 |
| SRMS | PLXNA4 | 0.599658 | 4 | 0 | 3 |
| SRMS | IQCJ-SCHIP1-AS1 | 0.599367 | 3 | 0 | 3 |
| SRMS | AGXT | 0.596083 | 3 | 0 | 3 |
| SRMS | SPI1 | 0.595006 | 3 | 0 | 3 |
| SRMS | HMCN2 | 0.592864 | 3 | 0 | 3 |
| SRMS | SGK2 | 0.592025 | 3 | 0 | 3 |
| SRMS | ARX | 0.589957 | 4 | 0 | 3 |
| SRMS | GRAMD1B | 0.586707 | 4 | 0 | 3 |
| SRMS | GTPBP2 | 0.582355 | 3 | 0 | 3 |
For details and further investigation, click here