| Full name: tyrosine aminotransferase | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 16q22.2 | ||
| Entrez ID: 6898 | HGNC ID: HGNC:11573 | Ensembl Gene: ENSG00000198650 | OMIM ID: 613018 |
| Drug and gene relationship at DGIdb | |||
Expression of TAT:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TAT | 6898 | 206916_x_at | 0.0026 | 0.9914 | |
| GSE20347 | TAT | 6898 | 206916_x_at | -0.0508 | 0.4909 | |
| GSE23400 | TAT | 6898 | 214413_at | -0.1053 | 0.0002 | |
| GSE26886 | TAT | 6898 | 206916_x_at | -0.1826 | 0.1241 | |
| GSE29001 | TAT | 6898 | 214413_at | -0.1593 | 0.1519 | |
| GSE38129 | TAT | 6898 | 206916_x_at | -0.1445 | 0.0089 | |
| GSE45670 | TAT | 6898 | 206916_x_at | 0.0266 | 0.7842 | |
| GSE63941 | TAT | 6898 | 206916_x_at | 0.4604 | 0.0009 | |
| GSE77861 | TAT | 6898 | 1555189_a_at | -0.1489 | 0.2241 | |
| SRP133303 | TAT | 6898 | RNAseq | 0.0428 | 0.8529 | |
| TCGA | TAT | 6898 | RNAseq | 2.6166 | 0.0213 |
Upregulated datasets: 1; Downregulated datasets: 0.
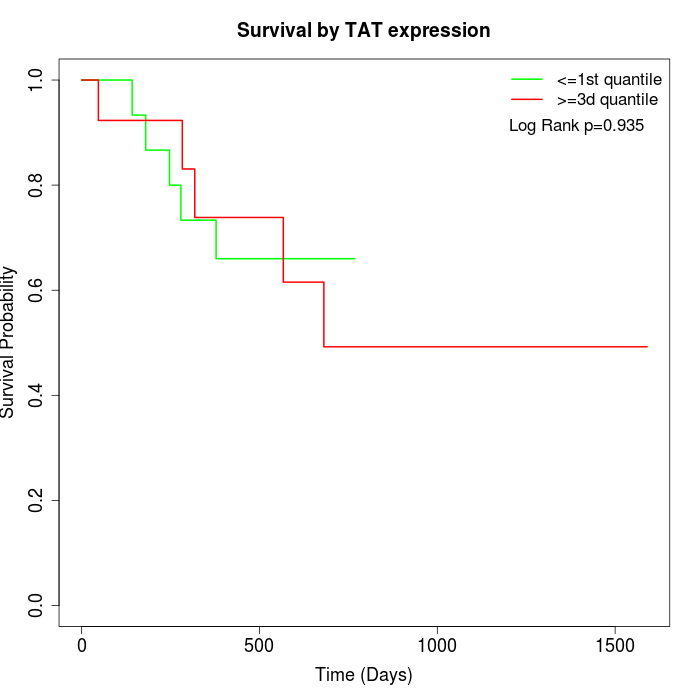
Survival by TAT expression:
Note: Click image to view full size file.
Copy number change of TAT:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TAT | 6898 | 4 | 0 | 26 | |
| GSE20123 | TAT | 6898 | 4 | 0 | 26 | |
| GSE43470 | TAT | 6898 | 2 | 8 | 33 | |
| GSE46452 | TAT | 6898 | 38 | 2 | 19 | |
| GSE47630 | TAT | 6898 | 11 | 8 | 21 | |
| GSE54993 | TAT | 6898 | 2 | 4 | 64 | |
| GSE54994 | TAT | 6898 | 8 | 10 | 35 | |
| GSE60625 | TAT | 6898 | 4 | 0 | 7 | |
| GSE74703 | TAT | 6898 | 2 | 5 | 29 | |
| GSE74704 | TAT | 6898 | 2 | 0 | 18 | |
| TCGA | TAT | 6898 | 28 | 12 | 56 |
Total number of gains: 105; Total number of losses: 49; Total Number of normals: 334.
Somatic mutations of TAT:
Generating mutation plots.
Highly correlated genes for TAT:
Showing top 20/508 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TAT | TCTN2 | 0.697154 | 3 | 0 | 3 |
| TAT | LIME1 | 0.692033 | 3 | 0 | 3 |
| TAT | DAND5 | 0.683844 | 3 | 0 | 3 |
| TAT | KRTAP4-12 | 0.664231 | 3 | 0 | 3 |
| TAT | NRGN | 0.661214 | 3 | 0 | 3 |
| TAT | BPIFB6 | 0.659158 | 3 | 0 | 3 |
| TAT | LINC00943 | 0.651806 | 4 | 0 | 4 |
| TAT | TSPO2 | 0.649691 | 6 | 0 | 6 |
| TAT | STAC | 0.645834 | 3 | 0 | 3 |
| TAT | KIAA0754 | 0.645816 | 3 | 0 | 3 |
| TAT | SPATA24 | 0.644672 | 5 | 0 | 5 |
| TAT | LINC01192 | 0.644454 | 3 | 0 | 3 |
| TAT | MATN1 | 0.642241 | 3 | 0 | 3 |
| TAT | PREX2 | 0.639632 | 3 | 0 | 3 |
| TAT | CLCNKB | 0.628882 | 3 | 0 | 3 |
| TAT | CRYBA4 | 0.628696 | 4 | 0 | 4 |
| TAT | LMAN1L | 0.625748 | 6 | 0 | 6 |
| TAT | MUC5AC | 0.625582 | 6 | 0 | 5 |
| TAT | SPX | 0.625581 | 4 | 0 | 3 |
| TAT | KLF14 | 0.624997 | 4 | 0 | 3 |
For details and further investigation, click here