| Full name: crystallin beta A4 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 22q12.1 | ||
| Entrez ID: 1413 | HGNC ID: HGNC:2396 | Ensembl Gene: ENSG00000196431 | OMIM ID: 123631 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of CRYBA4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CRYBA4 | 1413 | 206843_at | 0.0186 | 0.9599 | |
| GSE20347 | CRYBA4 | 1413 | 206843_at | -0.0588 | 0.4111 | |
| GSE23400 | CRYBA4 | 1413 | 206843_at | -0.1732 | 0.0000 | |
| GSE26886 | CRYBA4 | 1413 | 206843_at | 0.0969 | 0.4497 | |
| GSE29001 | CRYBA4 | 1413 | 206843_at | -0.1164 | 0.2730 | |
| GSE38129 | CRYBA4 | 1413 | 206843_at | -0.1388 | 0.0484 | |
| GSE45670 | CRYBA4 | 1413 | 206843_at | -0.0120 | 0.9378 | |
| GSE53622 | CRYBA4 | 1413 | 75728 | -0.0302 | 0.7434 | |
| GSE53624 | CRYBA4 | 1413 | 75728 | -0.0908 | 0.2421 | |
| GSE63941 | CRYBA4 | 1413 | 206843_at | 0.0368 | 0.8264 | |
| GSE77861 | CRYBA4 | 1413 | 206843_at | -0.1130 | 0.2520 |
Upregulated datasets: 0; Downregulated datasets: 0.
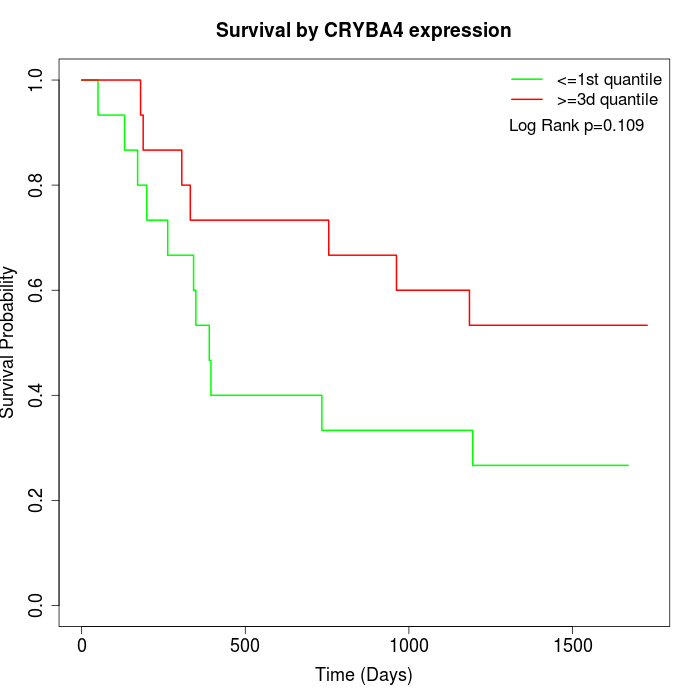
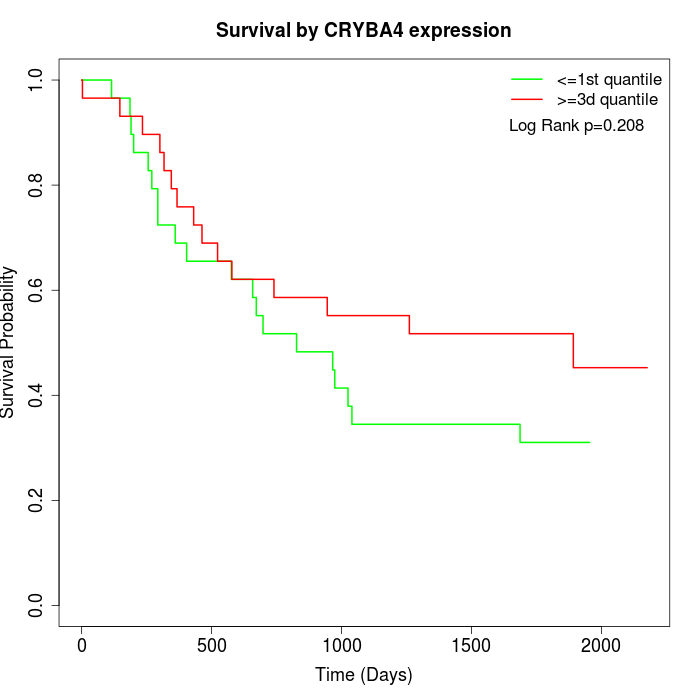
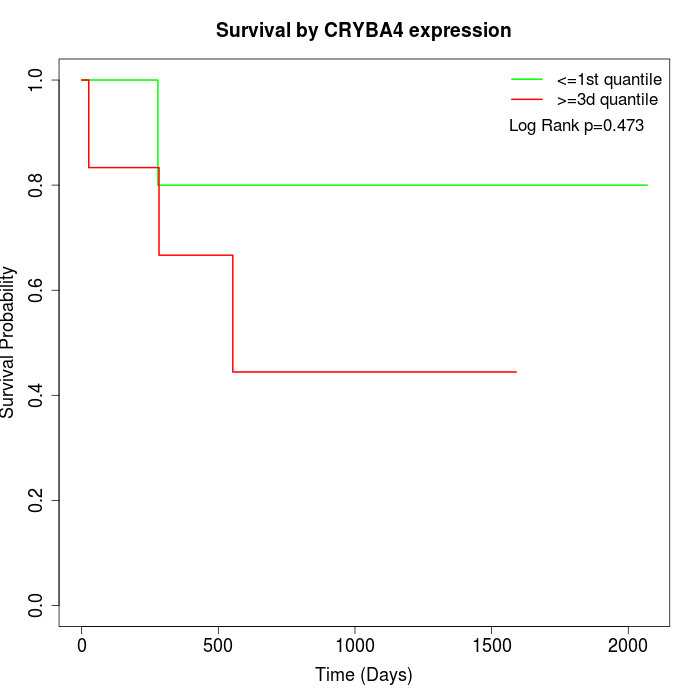
Survival by CRYBA4 expression:
Note: Click image to view full size file.
Copy number change of CRYBA4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CRYBA4 | 1413 | 3 | 5 | 22 | |
| GSE20123 | CRYBA4 | 1413 | 3 | 4 | 23 | |
| GSE43470 | CRYBA4 | 1413 | 4 | 6 | 33 | |
| GSE46452 | CRYBA4 | 1413 | 31 | 1 | 27 | |
| GSE47630 | CRYBA4 | 1413 | 8 | 5 | 27 | |
| GSE54993 | CRYBA4 | 1413 | 3 | 6 | 61 | |
| GSE54994 | CRYBA4 | 1413 | 11 | 8 | 34 | |
| GSE60625 | CRYBA4 | 1413 | 5 | 0 | 6 | |
| GSE74703 | CRYBA4 | 1413 | 3 | 5 | 28 | |
| GSE74704 | CRYBA4 | 1413 | 0 | 1 | 19 | |
| TCGA | CRYBA4 | 1413 | 26 | 14 | 56 |
Total number of gains: 97; Total number of losses: 55; Total Number of normals: 336.
Somatic mutations of CRYBA4:
Generating mutation plots.
Highly correlated genes for CRYBA4:
Showing top 20/565 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CRYBA4 | CLCNKB | 0.72808 | 4 | 0 | 4 |
| CRYBA4 | RNF17 | 0.683337 | 3 | 0 | 3 |
| CRYBA4 | NAT2 | 0.682371 | 3 | 0 | 3 |
| CRYBA4 | THRB | 0.68197 | 3 | 0 | 3 |
| CRYBA4 | TSPO2 | 0.681158 | 4 | 0 | 3 |
| CRYBA4 | PRODH2 | 0.669251 | 4 | 0 | 4 |
| CRYBA4 | RCVRN | 0.668279 | 4 | 0 | 3 |
| CRYBA4 | CST8 | 0.662408 | 5 | 0 | 5 |
| CRYBA4 | FGF6 | 0.660965 | 4 | 0 | 3 |
| CRYBA4 | HCG9 | 0.660004 | 4 | 0 | 4 |
| CRYBA4 | HRH3 | 0.656967 | 4 | 0 | 4 |
| CRYBA4 | OR2J2 | 0.656268 | 4 | 0 | 4 |
| CRYBA4 | RASL10A | 0.649307 | 3 | 0 | 3 |
| CRYBA4 | SCGN | 0.645763 | 3 | 0 | 3 |
| CRYBA4 | DKK4 | 0.644417 | 3 | 0 | 3 |
| CRYBA4 | SLC5A5 | 0.642288 | 4 | 0 | 3 |
| CRYBA4 | KSR1 | 0.640453 | 3 | 0 | 3 |
| CRYBA4 | MTAP | 0.640438 | 4 | 0 | 4 |
| CRYBA4 | LRTM1 | 0.640323 | 3 | 0 | 3 |
| CRYBA4 | C11orf16 | 0.639718 | 4 | 0 | 4 |
For details and further investigation, click here