| Full name: lectin, mannose binding 1 like | Alias Symbol: ERGL|ERGIC-53L | ||
| Type: protein-coding gene | Cytoband: 15q24.1 | ||
| Entrez ID: 79748 | HGNC ID: HGNC:6632 | Ensembl Gene: ENSG00000140506 | OMIM ID: 609548 |
| Drug and gene relationship at DGIdb | |||
Expression of LMAN1L:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LMAN1L | 79748 | 220420_at | -0.0923 | 0.6806 | |
| GSE20347 | LMAN1L | 79748 | 220420_at | -0.0164 | 0.8704 | |
| GSE23400 | LMAN1L | 79748 | 220420_at | -0.1681 | 0.0000 | |
| GSE26886 | LMAN1L | 79748 | 220420_at | -0.1970 | 0.1478 | |
| GSE29001 | LMAN1L | 79748 | 220420_at | -0.1438 | 0.4994 | |
| GSE38129 | LMAN1L | 79748 | 220420_at | -0.0916 | 0.1804 | |
| GSE45670 | LMAN1L | 79748 | 220420_at | -0.0375 | 0.6430 | |
| GSE63941 | LMAN1L | 79748 | 220420_at | 0.0049 | 0.9724 | |
| GSE77861 | LMAN1L | 79748 | 220420_at | -0.1201 | 0.1776 | |
| GSE97050 | LMAN1L | 79748 | A_23_P140630 | 0.1899 | 0.3286 | |
| TCGA | LMAN1L | 79748 | RNAseq | -0.7703 | 0.5385 |
Upregulated datasets: 0; Downregulated datasets: 0.
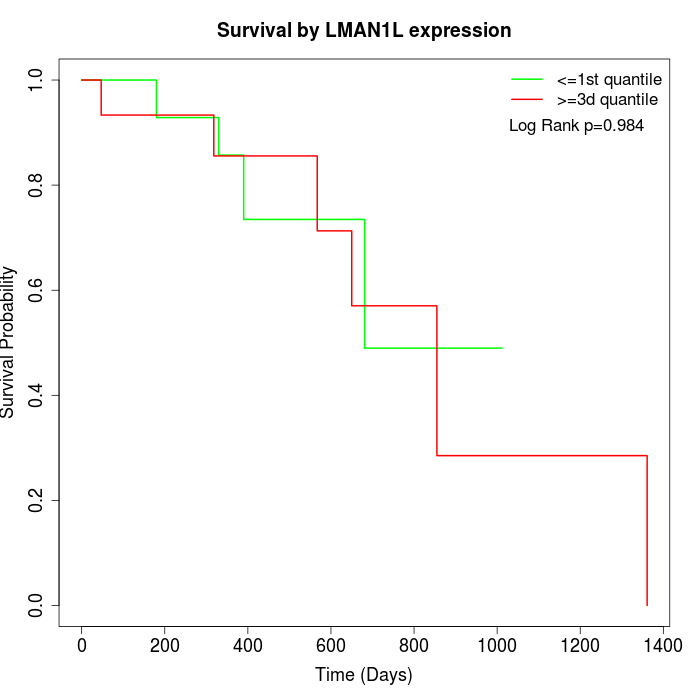
Survival by LMAN1L expression:
Note: Click image to view full size file.
Copy number change of LMAN1L:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LMAN1L | 79748 | 8 | 1 | 21 | |
| GSE20123 | LMAN1L | 79748 | 8 | 1 | 21 | |
| GSE43470 | LMAN1L | 79748 | 4 | 6 | 33 | |
| GSE46452 | LMAN1L | 79748 | 3 | 7 | 49 | |
| GSE47630 | LMAN1L | 79748 | 8 | 10 | 22 | |
| GSE54993 | LMAN1L | 79748 | 5 | 6 | 59 | |
| GSE54994 | LMAN1L | 79748 | 5 | 7 | 41 | |
| GSE60625 | LMAN1L | 79748 | 4 | 0 | 7 | |
| GSE74703 | LMAN1L | 79748 | 4 | 3 | 29 | |
| GSE74704 | LMAN1L | 79748 | 4 | 1 | 15 | |
| TCGA | LMAN1L | 79748 | 14 | 15 | 67 |
Total number of gains: 67; Total number of losses: 57; Total Number of normals: 364.
Somatic mutations of LMAN1L:
Generating mutation plots.
Highly correlated genes for LMAN1L:
Showing top 20/1373 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LMAN1L | LIMD1 | 0.809395 | 3 | 0 | 3 |
| LMAN1L | PTGDR2 | 0.788689 | 4 | 0 | 4 |
| LMAN1L | ERN1 | 0.782431 | 4 | 0 | 4 |
| LMAN1L | GRID2 | 0.771478 | 5 | 0 | 5 |
| LMAN1L | ZBTB45 | 0.768917 | 3 | 0 | 3 |
| LMAN1L | DAB1 | 0.767202 | 5 | 0 | 5 |
| LMAN1L | ABCC6 | 0.764293 | 4 | 0 | 4 |
| LMAN1L | VGF | 0.756333 | 3 | 0 | 3 |
| LMAN1L | SUN2 | 0.754835 | 3 | 0 | 3 |
| LMAN1L | WDR38 | 0.754765 | 3 | 0 | 3 |
| LMAN1L | C10orf90 | 0.753993 | 3 | 0 | 3 |
| LMAN1L | C16orf82 | 0.750775 | 3 | 0 | 3 |
| LMAN1L | DMP1 | 0.735078 | 3 | 0 | 3 |
| LMAN1L | ADM2 | 0.733216 | 8 | 0 | 8 |
| LMAN1L | BTBD19 | 0.732619 | 3 | 0 | 3 |
| LMAN1L | GALNT9 | 0.730334 | 3 | 0 | 3 |
| LMAN1L | CLDN9 | 0.729058 | 7 | 0 | 6 |
| LMAN1L | KCNA7 | 0.728943 | 3 | 0 | 3 |
| LMAN1L | AGRP | 0.728324 | 5 | 0 | 5 |
| LMAN1L | PSPN | 0.727234 | 6 | 0 | 5 |
For details and further investigation, click here