| Full name: transcription elongation factor A like 4 | Alias Symbol: FLJ21174|WEX7 | ||
| Type: protein-coding gene | Cytoband: Xq22.2 | ||
| Entrez ID: 79921 | HGNC ID: HGNC:26121 | Ensembl Gene: ENSG00000133142 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of TCEAL4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TCEAL4 | 79921 | 202371_at | -0.6892 | 0.1635 | |
| GSE20347 | TCEAL4 | 79921 | 202371_at | 0.0383 | 0.8331 | |
| GSE23400 | TCEAL4 | 79921 | 202371_at | 0.0461 | 0.5774 | |
| GSE26886 | TCEAL4 | 79921 | 202371_at | 0.9723 | 0.0000 | |
| GSE29001 | TCEAL4 | 79921 | 202371_at | 0.2207 | 0.3356 | |
| GSE38129 | TCEAL4 | 79921 | 202371_at | -0.1114 | 0.6105 | |
| GSE45670 | TCEAL4 | 79921 | 202371_at | -0.7007 | 0.0010 | |
| GSE53622 | TCEAL4 | 79921 | 121507 | -0.5012 | 0.0000 | |
| GSE53624 | TCEAL4 | 79921 | 121507 | -0.2807 | 0.0001 | |
| GSE63941 | TCEAL4 | 79921 | 202371_at | -0.6137 | 0.2091 | |
| GSE77861 | TCEAL4 | 79921 | 202371_at | -0.0627 | 0.9007 | |
| GSE97050 | TCEAL4 | 79921 | A_23_P259166 | -1.0663 | 0.0620 | |
| SRP007169 | TCEAL4 | 79921 | RNAseq | -0.1279 | 0.6845 | |
| SRP008496 | TCEAL4 | 79921 | RNAseq | 0.4536 | 0.1082 | |
| SRP064894 | TCEAL4 | 79921 | RNAseq | -0.4821 | 0.0279 | |
| SRP133303 | TCEAL4 | 79921 | RNAseq | -0.1438 | 0.5697 | |
| SRP159526 | TCEAL4 | 79921 | RNAseq | -0.1983 | 0.4520 | |
| SRP193095 | TCEAL4 | 79921 | RNAseq | -0.0159 | 0.9376 | |
| SRP219564 | TCEAL4 | 79921 | RNAseq | -0.6297 | 0.2135 | |
| TCGA | TCEAL4 | 79921 | RNAseq | -0.1180 | 0.0342 |
Upregulated datasets: 0; Downregulated datasets: 0.
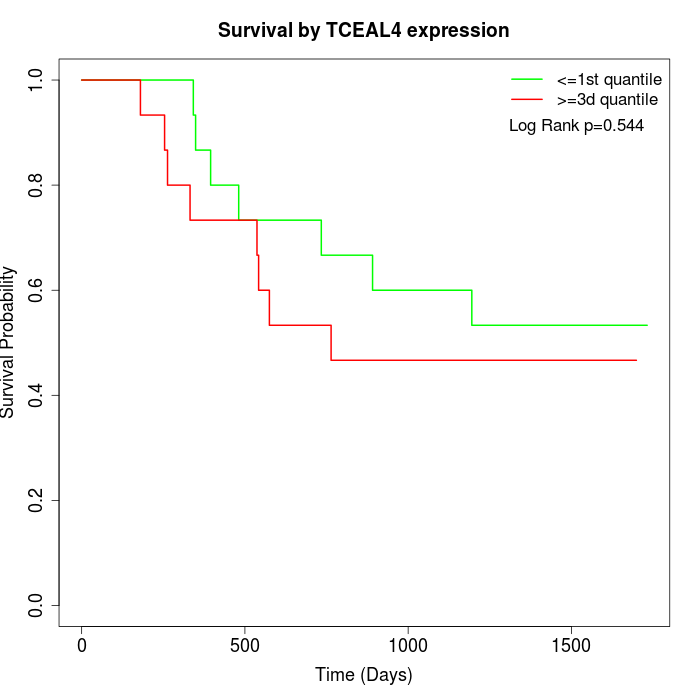
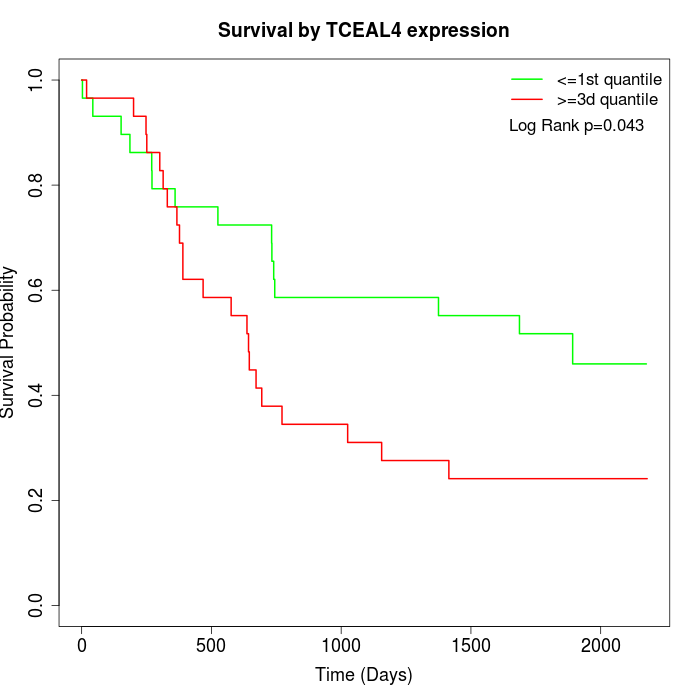
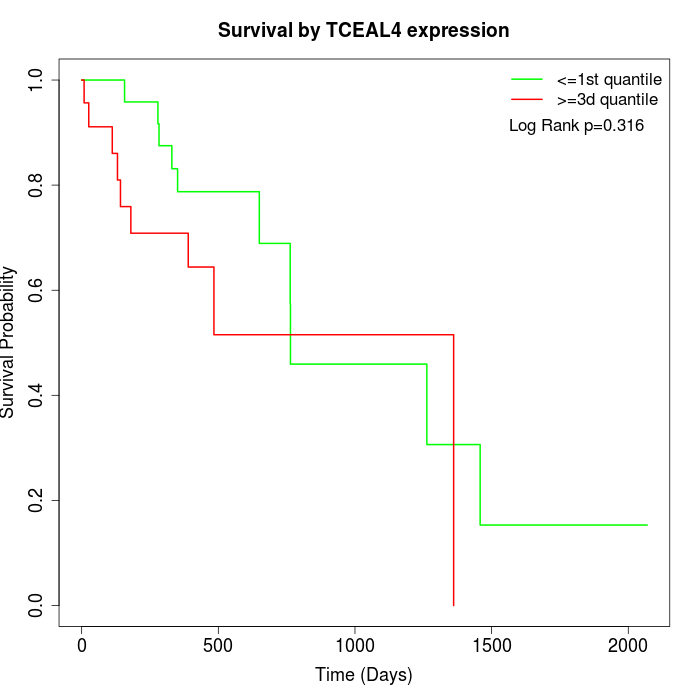
Survival by TCEAL4 expression:
Note: Click image to view full size file.
Copy number change of TCEAL4:
No record found for this gene.
Somatic mutations of TCEAL4:
Generating mutation plots.
Highly correlated genes for TCEAL4:
Showing top 20/697 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TCEAL4 | TCEAL5 | 0.788191 | 3 | 0 | 3 |
| TCEAL4 | TCEAL1 | 0.762928 | 11 | 0 | 11 |
| TCEAL4 | MYH3 | 0.751907 | 3 | 0 | 3 |
| TCEAL4 | UQCRFS1 | 0.745905 | 3 | 0 | 3 |
| TCEAL4 | ANKS1B | 0.744127 | 3 | 0 | 3 |
| TCEAL4 | ZBTB26 | 0.734329 | 4 | 0 | 4 |
| TCEAL4 | EIF5B | 0.716918 | 3 | 0 | 3 |
| TCEAL4 | TCEAL3 | 0.714218 | 6 | 0 | 5 |
| TCEAL4 | ZNF766 | 0.710979 | 3 | 0 | 3 |
| TCEAL4 | SATB1 | 0.705528 | 3 | 0 | 3 |
| TCEAL4 | ZCRB1 | 0.704975 | 3 | 0 | 3 |
| TCEAL4 | FBXL4 | 0.70443 | 3 | 0 | 3 |
| TCEAL4 | PNCK | 0.702783 | 3 | 0 | 3 |
| TCEAL4 | MAPK1IP1L | 0.69864 | 3 | 0 | 3 |
| TCEAL4 | ZXDA | 0.698395 | 3 | 0 | 3 |
| TCEAL4 | MOAP1 | 0.693809 | 12 | 0 | 10 |
| TCEAL4 | RNASEH2C | 0.684709 | 3 | 0 | 3 |
| TCEAL4 | RIMKLB | 0.684354 | 4 | 0 | 4 |
| TCEAL4 | INTU | 0.683907 | 3 | 0 | 3 |
| TCEAL4 | RNASE4 | 0.679067 | 6 | 0 | 5 |
For details and further investigation, click here