| Full name: transcription elongation factor A like 1 | Alias Symbol: p21|pp21|SIIR|P21|WEX9 | ||
| Type: protein-coding gene | Cytoband: Xq22.2 | ||
| Entrez ID: 9338 | HGNC ID: HGNC:11616 | Ensembl Gene: ENSG00000172465 | OMIM ID: 300237 |
| Drug and gene relationship at DGIdb | |||
Expression of TCEAL1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TCEAL1 | 9338 | 204045_at | -1.0541 | 0.0696 | |
| GSE20347 | TCEAL1 | 9338 | 204045_at | -0.0823 | 0.6757 | |
| GSE23400 | TCEAL1 | 9338 | 204045_at | -0.0947 | 0.2516 | |
| GSE26886 | TCEAL1 | 9338 | 204045_at | 0.6956 | 0.0360 | |
| GSE29001 | TCEAL1 | 9338 | 204045_at | 0.0650 | 0.8135 | |
| GSE38129 | TCEAL1 | 9338 | 204045_at | -0.4077 | 0.1307 | |
| GSE45670 | TCEAL1 | 9338 | 204045_at | -1.1721 | 0.0000 | |
| GSE53622 | TCEAL1 | 9338 | 52790 | -0.8492 | 0.0000 | |
| GSE53624 | TCEAL1 | 9338 | 52790 | -0.9303 | 0.0000 | |
| GSE63941 | TCEAL1 | 9338 | 204045_at | -0.6378 | 0.5067 | |
| GSE77861 | TCEAL1 | 9338 | 204045_at | 0.1895 | 0.4123 | |
| GSE97050 | TCEAL1 | 9338 | A_23_P73801 | -0.7432 | 0.1450 | |
| SRP007169 | TCEAL1 | 9338 | RNAseq | -0.2693 | 0.5506 | |
| SRP008496 | TCEAL1 | 9338 | RNAseq | 0.6313 | 0.1059 | |
| SRP064894 | TCEAL1 | 9338 | RNAseq | -0.8476 | 0.0005 | |
| SRP133303 | TCEAL1 | 9338 | RNAseq | -0.4805 | 0.0758 | |
| SRP159526 | TCEAL1 | 9338 | RNAseq | -0.6384 | 0.0497 | |
| SRP193095 | TCEAL1 | 9338 | RNAseq | -0.1365 | 0.3826 | |
| SRP219564 | TCEAL1 | 9338 | RNAseq | -0.8046 | 0.1612 | |
| TCGA | TCEAL1 | 9338 | RNAseq | -0.3597 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
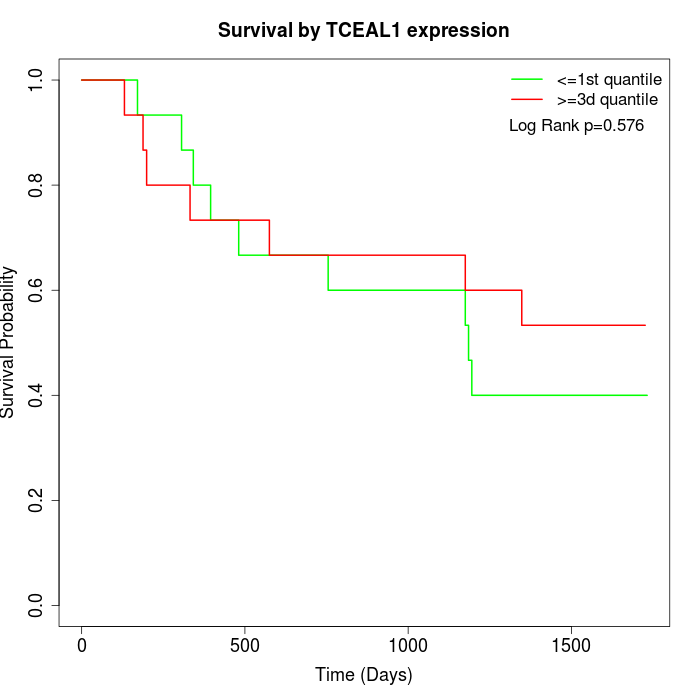
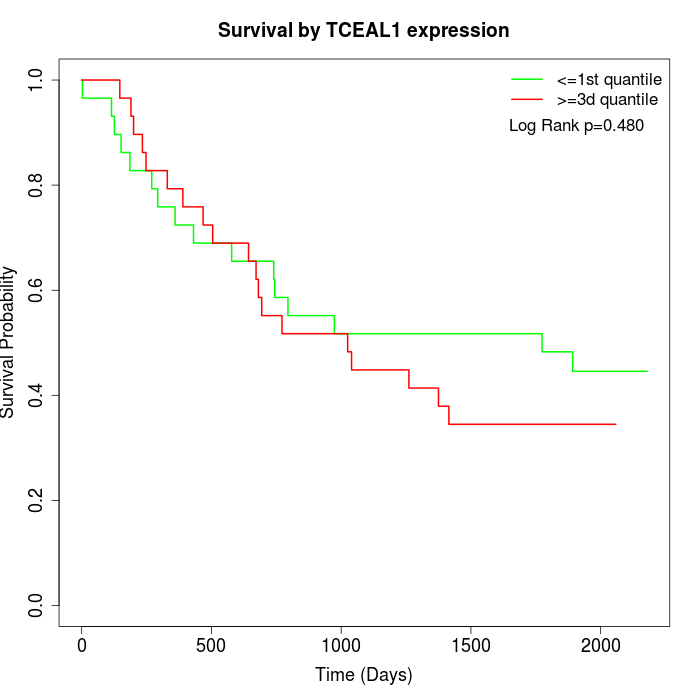
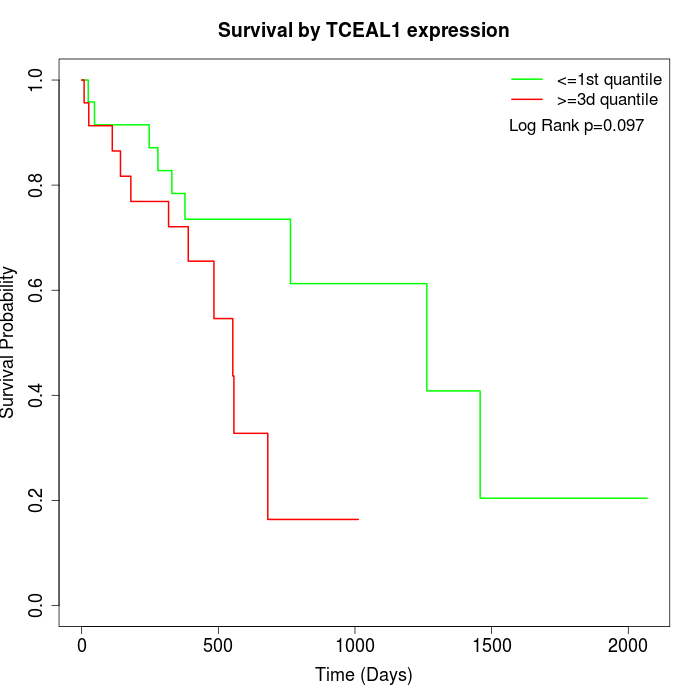
Survival by TCEAL1 expression:
Note: Click image to view full size file.
Copy number change of TCEAL1:
No record found for this gene.
Somatic mutations of TCEAL1:
Generating mutation plots.
Highly correlated genes for TCEAL1:
Showing top 20/1055 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TCEAL1 | NFATC4 | 0.831512 | 3 | 0 | 3 |
| TCEAL1 | CBX7 | 0.802491 | 7 | 0 | 7 |
| TCEAL1 | NEGR1 | 0.801872 | 5 | 0 | 5 |
| TCEAL1 | C14orf28 | 0.799883 | 4 | 0 | 4 |
| TCEAL1 | RBPMS2 | 0.796868 | 5 | 0 | 5 |
| TCEAL1 | LRRN4CL | 0.793329 | 5 | 0 | 5 |
| TCEAL1 | LRFN5 | 0.792381 | 4 | 0 | 4 |
| TCEAL1 | WDR48 | 0.784868 | 3 | 0 | 3 |
| TCEAL1 | FBXL3 | 0.779276 | 3 | 0 | 3 |
| TCEAL1 | CLEC3B | 0.777016 | 3 | 0 | 3 |
| TCEAL1 | EID3 | 0.773419 | 3 | 0 | 3 |
| TCEAL1 | TCEAL5 | 0.772238 | 3 | 0 | 3 |
| TCEAL1 | ANO5 | 0.772075 | 5 | 0 | 5 |
| TCEAL1 | MOAP1 | 0.769546 | 11 | 0 | 11 |
| TCEAL1 | MRVI1 | 0.767416 | 5 | 0 | 5 |
| TCEAL1 | CASQ2 | 0.763692 | 8 | 0 | 8 |
| TCEAL1 | TCEAL4 | 0.762928 | 11 | 0 | 11 |
| TCEAL1 | FDX1 | 0.760022 | 3 | 0 | 3 |
| TCEAL1 | KLHL15 | 0.75904 | 5 | 0 | 5 |
| TCEAL1 | MECP2 | 0.755759 | 4 | 0 | 4 |
For details and further investigation, click here