| Full name: tripartite motif containing 15 | Alias Symbol: ZNFB7|RNF93 | ||
| Type: protein-coding gene | Cytoband: 6p22.1 | ||
| Entrez ID: 89870 | HGNC ID: HGNC:16284 | Ensembl Gene: ENSG00000204610 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of TRIM15:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TRIM15 | 89870 | 210177_at | -0.2232 | 0.2371 | |
| GSE20347 | TRIM15 | 89870 | 210177_at | -0.1874 | 0.0071 | |
| GSE23400 | TRIM15 | 89870 | 210177_at | -0.2094 | 0.0000 | |
| GSE26886 | TRIM15 | 89870 | 210177_at | -0.0457 | 0.7032 | |
| GSE29001 | TRIM15 | 89870 | 210177_at | -0.4073 | 0.0206 | |
| GSE38129 | TRIM15 | 89870 | 210177_at | -0.1399 | 0.0361 | |
| GSE45670 | TRIM15 | 89870 | 210177_at | -0.1546 | 0.0933 | |
| GSE53622 | TRIM15 | 89870 | 1299 | 0.4958 | 0.0004 | |
| GSE53624 | TRIM15 | 89870 | 1299 | 0.4921 | 0.0003 | |
| GSE63941 | TRIM15 | 89870 | 210177_at | 0.3115 | 0.2034 | |
| GSE77861 | TRIM15 | 89870 | 210177_at | -0.3386 | 0.0640 | |
| GSE97050 | TRIM15 | 89870 | A_23_P500300 | -0.0292 | 0.8966 | |
| TCGA | TRIM15 | 89870 | RNAseq | 0.5443 | 0.5033 |
Upregulated datasets: 0; Downregulated datasets: 0.
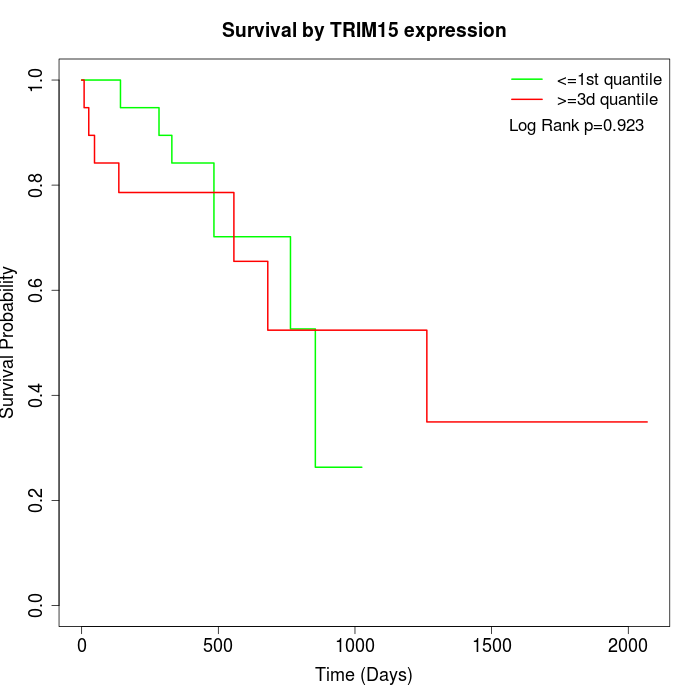
Survival by TRIM15 expression:
Note: Click image to view full size file.
Copy number change of TRIM15:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRIM15 | 89870 | 4 | 1 | 25 | |
| GSE20123 | TRIM15 | 89870 | 4 | 1 | 25 | |
| GSE43470 | TRIM15 | 89870 | 5 | 0 | 38 | |
| GSE46452 | TRIM15 | 89870 | 1 | 10 | 48 | |
| GSE47630 | TRIM15 | 89870 | 7 | 3 | 30 | |
| GSE54993 | TRIM15 | 89870 | 2 | 1 | 67 | |
| GSE54994 | TRIM15 | 89870 | 11 | 4 | 38 | |
| GSE60625 | TRIM15 | 89870 | 0 | 1 | 10 | |
| GSE74703 | TRIM15 | 89870 | 5 | 0 | 31 | |
| GSE74704 | TRIM15 | 89870 | 2 | 0 | 18 | |
| TCGA | TRIM15 | 89870 | 16 | 17 | 63 |
Total number of gains: 57; Total number of losses: 38; Total Number of normals: 393.
Somatic mutations of TRIM15:
Generating mutation plots.
Highly correlated genes for TRIM15:
Showing top 20/565 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRIM15 | KRTAP10-9 | 0.719084 | 3 | 0 | 3 |
| TRIM15 | NPFF | 0.703394 | 3 | 0 | 3 |
| TRIM15 | CNR1 | 0.684733 | 4 | 0 | 4 |
| TRIM15 | SH2B1 | 0.682986 | 3 | 0 | 3 |
| TRIM15 | PCBP3 | 0.679742 | 3 | 0 | 3 |
| TRIM15 | CPEB4 | 0.679739 | 4 | 0 | 4 |
| TRIM15 | RBP2 | 0.67401 | 3 | 0 | 3 |
| TRIM15 | GUCA2B | 0.673156 | 3 | 0 | 3 |
| TRIM15 | ATG9B | 0.672219 | 3 | 0 | 3 |
| TRIM15 | ALK | 0.671911 | 6 | 0 | 5 |
| TRIM15 | FAM110D | 0.671712 | 5 | 0 | 5 |
| TRIM15 | SPEG | 0.67073 | 3 | 0 | 3 |
| TRIM15 | SNORA68 | 0.669704 | 3 | 0 | 3 |
| TRIM15 | TNFSF9 | 0.664895 | 3 | 0 | 3 |
| TRIM15 | GJC2 | 0.664625 | 8 | 0 | 7 |
| TRIM15 | RGS14 | 0.66399 | 7 | 0 | 7 |
| TRIM15 | STBD1 | 0.658446 | 4 | 0 | 4 |
| TRIM15 | NFAM1 | 0.656969 | 3 | 0 | 3 |
| TRIM15 | DUX1 | 0.64801 | 7 | 0 | 6 |
| TRIM15 | KAT7 | 0.64688 | 3 | 0 | 3 |
For details and further investigation, click here