| Full name: SH2B adaptor protein 1 | Alias Symbol: FLJ30542|SH2B | ||
| Type: protein-coding gene | Cytoband: 16p11.2 | ||
| Entrez ID: 25970 | HGNC ID: HGNC:30417 | Ensembl Gene: ENSG00000178188 | OMIM ID: 608937 |
| Drug and gene relationship at DGIdb | |||
SH2B1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04722 | Neurotrophin signaling pathway |
Expression of SH2B1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SH2B1 | 25970 | 40149_at | -0.0697 | 0.9076 | |
| GSE20347 | SH2B1 | 25970 | 209322_s_at | -0.0416 | 0.6290 | |
| GSE23400 | SH2B1 | 25970 | 40149_at | -0.0775 | 0.0236 | |
| GSE26886 | SH2B1 | 25970 | 209322_s_at | 0.3566 | 0.0083 | |
| GSE29001 | SH2B1 | 25970 | 40149_at | -0.1693 | 0.3616 | |
| GSE38129 | SH2B1 | 25970 | 40149_at | 0.0062 | 0.9572 | |
| GSE45670 | SH2B1 | 25970 | 40149_at | -0.1283 | 0.2770 | |
| GSE53622 | SH2B1 | 25970 | 19009 | -0.0381 | 0.6850 | |
| GSE53624 | SH2B1 | 25970 | 19009 | 0.2695 | 0.0002 | |
| GSE63941 | SH2B1 | 25970 | 209322_s_at | -0.2422 | 0.2607 | |
| GSE77861 | SH2B1 | 25970 | 209322_s_at | -0.1988 | 0.1455 | |
| GSE97050 | SH2B1 | 25970 | A_24_P185029 | -0.1602 | 0.5574 | |
| SRP007169 | SH2B1 | 25970 | RNAseq | -0.1385 | 0.7279 | |
| SRP008496 | SH2B1 | 25970 | RNAseq | 0.0350 | 0.9070 | |
| SRP064894 | SH2B1 | 25970 | RNAseq | 0.0453 | 0.8252 | |
| SRP133303 | SH2B1 | 25970 | RNAseq | -0.0715 | 0.6708 | |
| SRP159526 | SH2B1 | 25970 | RNAseq | 0.0729 | 0.8393 | |
| SRP193095 | SH2B1 | 25970 | RNAseq | 0.0418 | 0.7441 | |
| SRP219564 | SH2B1 | 25970 | RNAseq | -0.1532 | 0.7021 | |
| TCGA | SH2B1 | 25970 | RNAseq | -0.1927 | 0.0001 |
Upregulated datasets: 0; Downregulated datasets: 0.
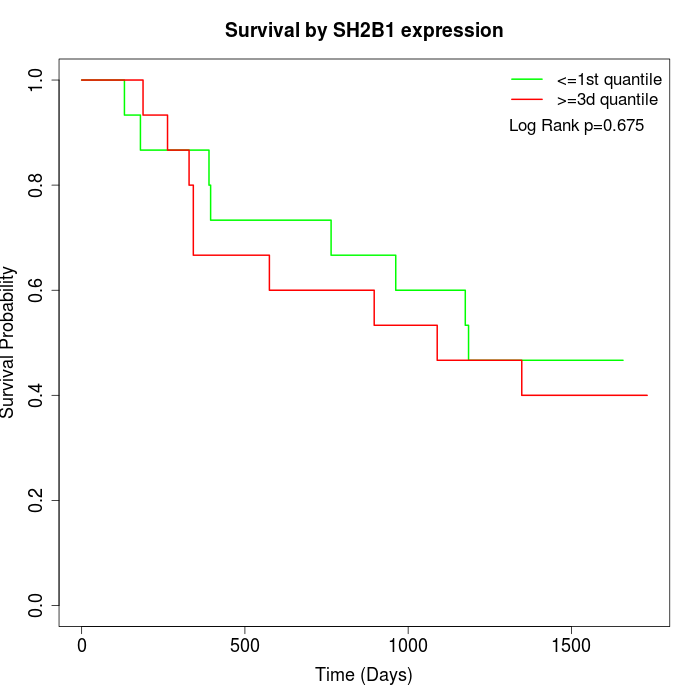
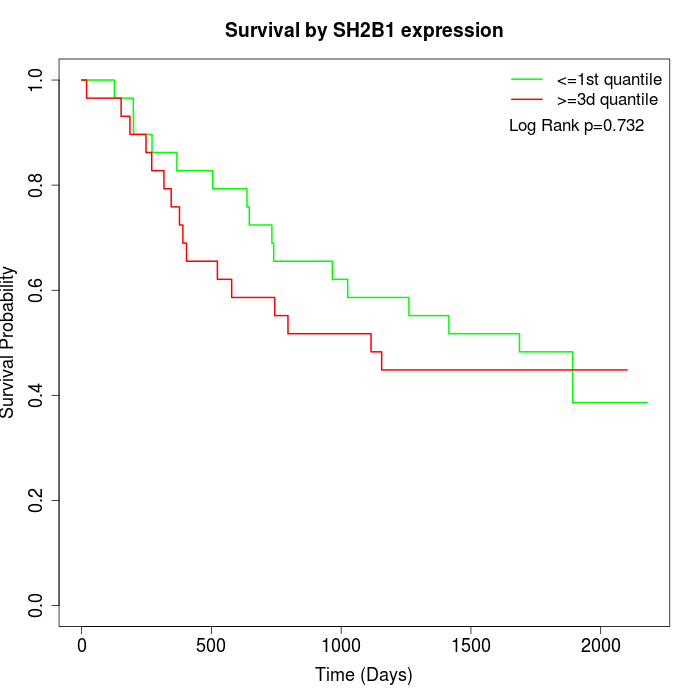
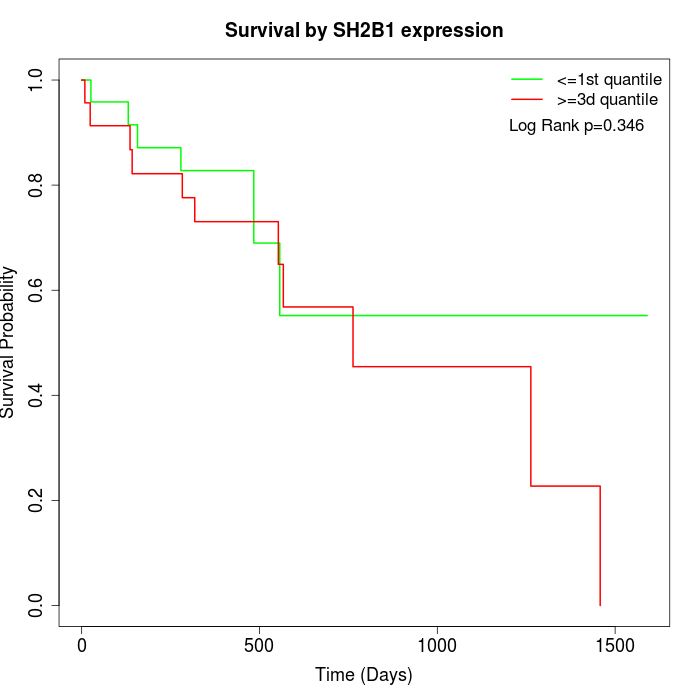
Survival by SH2B1 expression:
Note: Click image to view full size file.
Copy number change of SH2B1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SH2B1 | 25970 | 5 | 5 | 20 | |
| GSE20123 | SH2B1 | 25970 | 5 | 4 | 21 | |
| GSE43470 | SH2B1 | 25970 | 3 | 3 | 37 | |
| GSE46452 | SH2B1 | 25970 | 38 | 1 | 20 | |
| GSE47630 | SH2B1 | 25970 | 11 | 7 | 22 | |
| GSE54993 | SH2B1 | 25970 | 3 | 5 | 62 | |
| GSE54994 | SH2B1 | 25970 | 5 | 9 | 39 | |
| GSE60625 | SH2B1 | 25970 | 4 | 0 | 7 | |
| GSE74703 | SH2B1 | 25970 | 3 | 2 | 31 | |
| GSE74704 | SH2B1 | 25970 | 3 | 2 | 15 | |
| TCGA | SH2B1 | 25970 | 20 | 12 | 64 |
Total number of gains: 100; Total number of losses: 50; Total Number of normals: 338.
Somatic mutations of SH2B1:
Generating mutation plots.
Highly correlated genes for SH2B1:
Showing top 20/507 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SH2B1 | NDUFB11 | 0.811829 | 3 | 0 | 3 |
| SH2B1 | ALKBH7 | 0.809545 | 3 | 0 | 3 |
| SH2B1 | MED26 | 0.777359 | 3 | 0 | 3 |
| SH2B1 | ZNF653 | 0.775984 | 4 | 0 | 4 |
| SH2B1 | BLOC1S1 | 0.775374 | 3 | 0 | 3 |
| SH2B1 | AKR7A2 | 0.7722 | 3 | 0 | 3 |
| SH2B1 | CS | 0.76063 | 3 | 0 | 3 |
| SH2B1 | FOXP4 | 0.759975 | 3 | 0 | 3 |
| SH2B1 | TAB1 | 0.758647 | 3 | 0 | 3 |
| SH2B1 | SPEN | 0.756075 | 3 | 0 | 3 |
| SH2B1 | SIN3A | 0.749366 | 3 | 0 | 3 |
| SH2B1 | R3HDM2 | 0.740902 | 3 | 0 | 3 |
| SH2B1 | RNFT2 | 0.737758 | 3 | 0 | 3 |
| SH2B1 | TSNARE1 | 0.737084 | 5 | 0 | 5 |
| SH2B1 | MATN4 | 0.72995 | 4 | 0 | 4 |
| SH2B1 | TBC1D25 | 0.725243 | 4 | 0 | 4 |
| SH2B1 | NCKIPSD | 0.723936 | 3 | 0 | 3 |
| SH2B1 | SELPLG | 0.723522 | 3 | 0 | 3 |
| SH2B1 | SNAPC2 | 0.717052 | 3 | 0 | 3 |
| SH2B1 | HK1 | 0.716151 | 3 | 0 | 3 |
For details and further investigation, click here