| Full name: guanylate cyclase activator 2B | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 1p34.2 | ||
| Entrez ID: 2981 | HGNC ID: HGNC:4683 | Ensembl Gene: ENSG00000044012 | OMIM ID: 601271 |
| Drug and gene relationship at DGIdb | |||
Expression of GUCA2B:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | GUCA2B | 2981 | 207502_at | -0.0403 | 0.8954 | |
| GSE20347 | GUCA2B | 2981 | 207502_at | -0.0133 | 0.8865 | |
| GSE23400 | GUCA2B | 2981 | 207502_at | -0.0573 | 0.0482 | |
| GSE26886 | GUCA2B | 2981 | 207502_at | -0.0213 | 0.8407 | |
| GSE29001 | GUCA2B | 2981 | 207502_at | -0.0231 | 0.8973 | |
| GSE38129 | GUCA2B | 2981 | 207502_at | -0.1291 | 0.0613 | |
| GSE45670 | GUCA2B | 2981 | 207502_at | 0.1533 | 0.1054 | |
| GSE53622 | GUCA2B | 2981 | 90846 | 0.1051 | 0.7025 | |
| GSE53624 | GUCA2B | 2981 | 90846 | -0.0868 | 0.6995 | |
| GSE63941 | GUCA2B | 2981 | 207502_at | 0.0577 | 0.8471 | |
| GSE77861 | GUCA2B | 2981 | 207502_at | -0.2268 | 0.0766 | |
| GSE97050 | GUCA2B | 2981 | A_23_P63032 | 0.0011 | 0.9975 | |
| TCGA | GUCA2B | 2981 | RNAseq | -7.0479 | 0.0001 |
Upregulated datasets: 0; Downregulated datasets: 1.
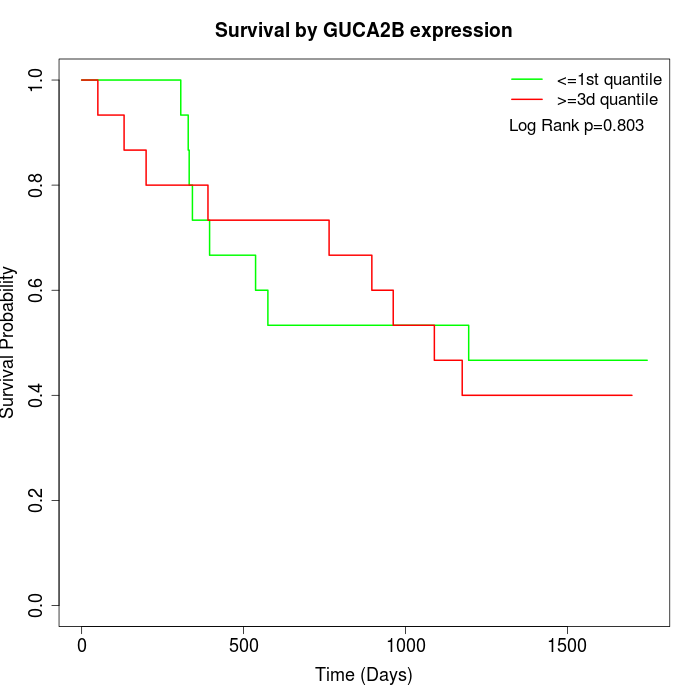
Survival by GUCA2B expression:
Note: Click image to view full size file.
Copy number change of GUCA2B:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | GUCA2B | 2981 | 3 | 4 | 23 | |
| GSE20123 | GUCA2B | 2981 | 3 | 2 | 25 | |
| GSE43470 | GUCA2B | 2981 | 7 | 2 | 34 | |
| GSE46452 | GUCA2B | 2981 | 3 | 1 | 55 | |
| GSE47630 | GUCA2B | 2981 | 9 | 2 | 29 | |
| GSE54993 | GUCA2B | 2981 | 0 | 2 | 68 | |
| GSE54994 | GUCA2B | 2981 | 13 | 2 | 38 | |
| GSE60625 | GUCA2B | 2981 | 0 | 0 | 11 | |
| GSE74703 | GUCA2B | 2981 | 6 | 1 | 29 | |
| GSE74704 | GUCA2B | 2981 | 1 | 0 | 19 | |
| TCGA | GUCA2B | 2981 | 13 | 16 | 67 |
Total number of gains: 58; Total number of losses: 32; Total Number of normals: 398.
Somatic mutations of GUCA2B:
Generating mutation plots.
Highly correlated genes for GUCA2B:
Showing top 20/702 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| GUCA2B | C20orf202 | 0.714257 | 3 | 0 | 3 |
| GUCA2B | VASH1 | 0.713451 | 3 | 0 | 3 |
| GUCA2B | LONRF3 | 0.709719 | 3 | 0 | 3 |
| GUCA2B | OR10H3 | 0.708353 | 4 | 0 | 4 |
| GUCA2B | ADAMTS18 | 0.705274 | 3 | 0 | 3 |
| GUCA2B | VGLL2 | 0.700372 | 3 | 0 | 3 |
| GUCA2B | ADAMTS12 | 0.694841 | 4 | 0 | 4 |
| GUCA2B | PDYN | 0.690489 | 6 | 0 | 6 |
| GUCA2B | BCAN | 0.689657 | 4 | 0 | 4 |
| GUCA2B | USP9Y | 0.687815 | 3 | 0 | 3 |
| GUCA2B | ATG10 | 0.68681 | 5 | 0 | 5 |
| GUCA2B | CHRM2 | 0.686774 | 5 | 0 | 5 |
| GUCA2B | SLITRK3 | 0.684824 | 3 | 0 | 3 |
| GUCA2B | HHLA1 | 0.683958 | 5 | 0 | 5 |
| GUCA2B | KCNN1 | 0.683493 | 6 | 0 | 6 |
| GUCA2B | KCND3 | 0.680522 | 4 | 0 | 3 |
| GUCA2B | CNTNAP1 | 0.678591 | 5 | 0 | 4 |
| GUCA2B | GRIK2 | 0.674104 | 3 | 0 | 3 |
| GUCA2B | TRIM15 | 0.673156 | 3 | 0 | 3 |
| GUCA2B | PEAK1 | 0.671117 | 6 | 0 | 5 |
For details and further investigation, click here