| Full name: tripartite motif containing 38 | Alias Symbol: RORET | ||
| Type: protein-coding gene | Cytoband: 6p22.2 | ||
| Entrez ID: 10475 | HGNC ID: HGNC:10059 | Ensembl Gene: ENSG00000112343 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of TRIM38:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TRIM38 | 10475 | 203568_s_at | -0.0537 | 0.9352 | |
| GSE20347 | TRIM38 | 10475 | 203568_s_at | -0.3395 | 0.0807 | |
| GSE23400 | TRIM38 | 10475 | 203567_s_at | -0.3140 | 0.0001 | |
| GSE26886 | TRIM38 | 10475 | 203567_s_at | -1.0116 | 0.0003 | |
| GSE29001 | TRIM38 | 10475 | 203567_s_at | -0.3319 | 0.2269 | |
| GSE38129 | TRIM38 | 10475 | 203568_s_at | -0.2447 | 0.0898 | |
| GSE45670 | TRIM38 | 10475 | 203567_s_at | -0.0366 | 0.9025 | |
| GSE53622 | TRIM38 | 10475 | 89914 | -0.3101 | 0.0004 | |
| GSE53624 | TRIM38 | 10475 | 89914 | -0.3742 | 0.0000 | |
| GSE63941 | TRIM38 | 10475 | 203568_s_at | -1.0825 | 0.1959 | |
| GSE77861 | TRIM38 | 10475 | 235084_x_at | 0.2610 | 0.4363 | |
| GSE97050 | TRIM38 | 10475 | A_24_P294851 | -0.1369 | 0.6950 | |
| SRP007169 | TRIM38 | 10475 | RNAseq | -0.0994 | 0.7743 | |
| SRP008496 | TRIM38 | 10475 | RNAseq | -0.4298 | 0.0429 | |
| SRP064894 | TRIM38 | 10475 | RNAseq | -0.3318 | 0.0530 | |
| SRP133303 | TRIM38 | 10475 | RNAseq | -0.4012 | 0.0053 | |
| SRP159526 | TRIM38 | 10475 | RNAseq | -0.6875 | 0.0603 | |
| SRP193095 | TRIM38 | 10475 | RNAseq | -0.3865 | 0.0033 | |
| SRP219564 | TRIM38 | 10475 | RNAseq | -0.5118 | 0.3140 | |
| TCGA | TRIM38 | 10475 | RNAseq | -0.0122 | 0.8620 |
Upregulated datasets: 0; Downregulated datasets: 1.
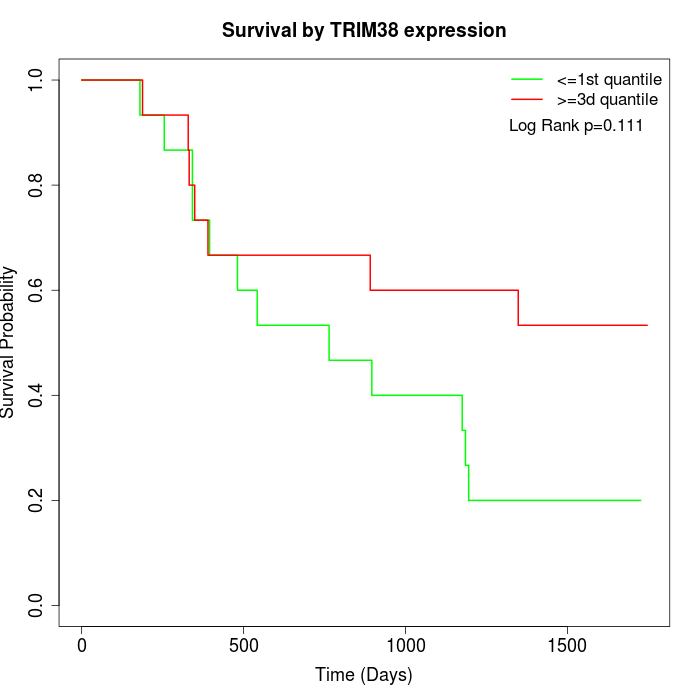
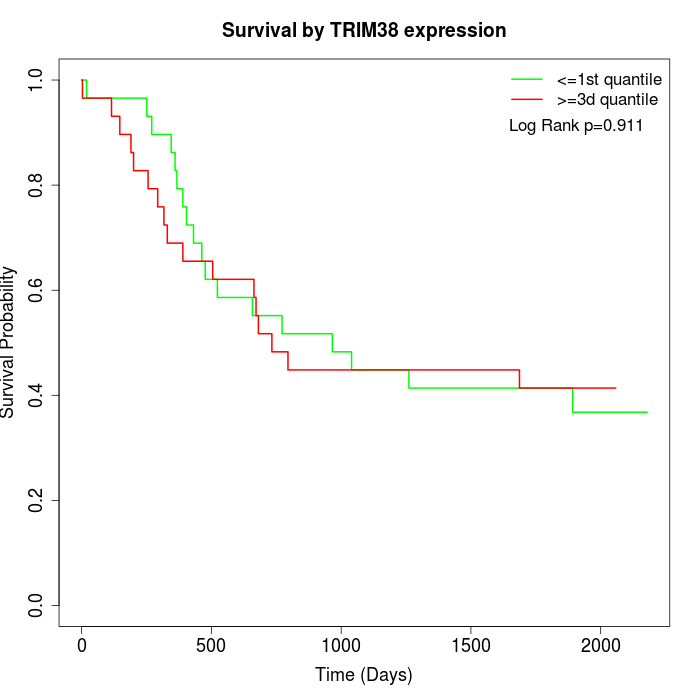
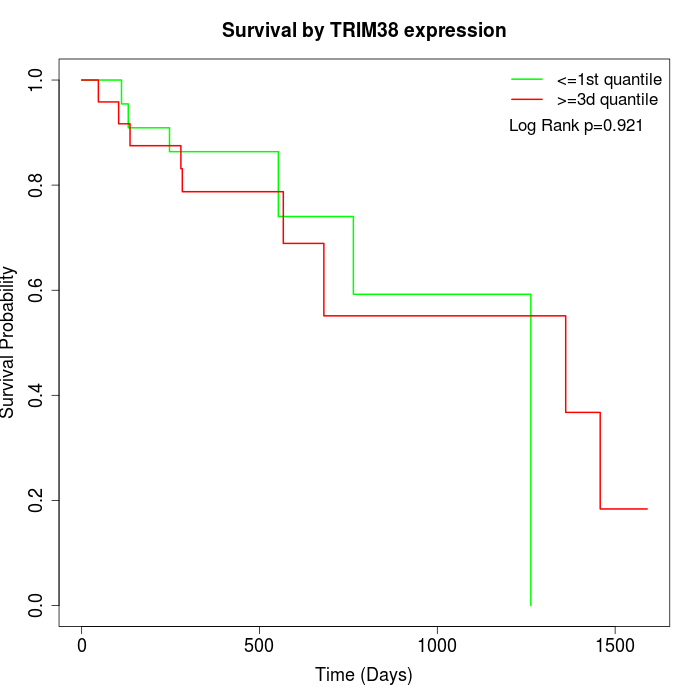
Survival by TRIM38 expression:
Note: Click image to view full size file.
Copy number change of TRIM38:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRIM38 | 10475 | 4 | 2 | 24 | |
| GSE20123 | TRIM38 | 10475 | 4 | 2 | 24 | |
| GSE43470 | TRIM38 | 10475 | 5 | 0 | 38 | |
| GSE46452 | TRIM38 | 10475 | 1 | 9 | 49 | |
| GSE47630 | TRIM38 | 10475 | 8 | 5 | 27 | |
| GSE54993 | TRIM38 | 10475 | 1 | 1 | 68 | |
| GSE54994 | TRIM38 | 10475 | 10 | 4 | 39 | |
| GSE60625 | TRIM38 | 10475 | 0 | 1 | 10 | |
| GSE74703 | TRIM38 | 10475 | 5 | 0 | 31 | |
| GSE74704 | TRIM38 | 10475 | 0 | 1 | 19 | |
| TCGA | TRIM38 | 10475 | 14 | 21 | 61 |
Total number of gains: 52; Total number of losses: 46; Total Number of normals: 390.
Somatic mutations of TRIM38:
Generating mutation plots.
Highly correlated genes for TRIM38:
Showing top 20/182 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRIM38 | ITPRIPL2 | 0.738876 | 3 | 0 | 3 |
| TRIM38 | CDK5RAP3 | 0.724695 | 3 | 0 | 3 |
| TRIM38 | PUS7L | 0.724222 | 3 | 0 | 3 |
| TRIM38 | ZNF681 | 0.722969 | 3 | 0 | 3 |
| TRIM38 | ZNF92 | 0.718359 | 3 | 0 | 3 |
| TRIM38 | ZNF256 | 0.713039 | 3 | 0 | 3 |
| TRIM38 | GDAP2 | 0.708048 | 3 | 0 | 3 |
| TRIM38 | ENPP5 | 0.705434 | 3 | 0 | 3 |
| TRIM38 | ZNF514 | 0.702979 | 3 | 0 | 3 |
| TRIM38 | SENP6 | 0.702791 | 3 | 0 | 3 |
| TRIM38 | ZNF800 | 0.692474 | 3 | 0 | 3 |
| TRIM38 | CCNYL1 | 0.690394 | 3 | 0 | 3 |
| TRIM38 | SMARCA4 | 0.688538 | 3 | 0 | 3 |
| TRIM38 | NAT1 | 0.687055 | 3 | 0 | 3 |
| TRIM38 | TTC17 | 0.680135 | 4 | 0 | 3 |
| TRIM38 | TAF9B | 0.675284 | 3 | 0 | 3 |
| TRIM38 | ZNF195 | 0.675249 | 3 | 0 | 3 |
| TRIM38 | GALNT11 | 0.666907 | 3 | 0 | 3 |
| TRIM38 | ZNF75A | 0.661295 | 4 | 0 | 3 |
| TRIM38 | SYTL2 | 0.659603 | 3 | 0 | 3 |
For details and further investigation, click here