| Full name: tetraspanin 16 | Alias Symbol: TM4-B|TM-8 | ||
| Type: protein-coding gene | Cytoband: 19p13.2 | ||
| Entrez ID: 26526 | HGNC ID: HGNC:30725 | Ensembl Gene: ENSG00000130167 | OMIM ID: 617580 |
| Drug and gene relationship at DGIdb | |||
Expression of TSPAN16:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TSPAN16 | 26526 | 233236_at | -0.0557 | 0.8245 | |
| GSE26886 | TSPAN16 | 26526 | 233236_at | -0.0154 | 0.9256 | |
| GSE45670 | TSPAN16 | 26526 | 233236_at | -0.0627 | 0.4109 | |
| GSE53622 | TSPAN16 | 26526 | 39875 | 0.3102 | 0.0007 | |
| GSE53624 | TSPAN16 | 26526 | 39875 | 0.1234 | 0.0485 | |
| GSE63941 | TSPAN16 | 26526 | 233236_at | 0.0615 | 0.6931 | |
| GSE77861 | TSPAN16 | 26526 | 233236_at | -0.1552 | 0.2723 | |
| SRP133303 | TSPAN16 | 26526 | RNAseq | -1.0687 | 0.0025 | |
| TCGA | TSPAN16 | 26526 | RNAseq | -1.6288 | 0.0965 |
Upregulated datasets: 0; Downregulated datasets: 1.
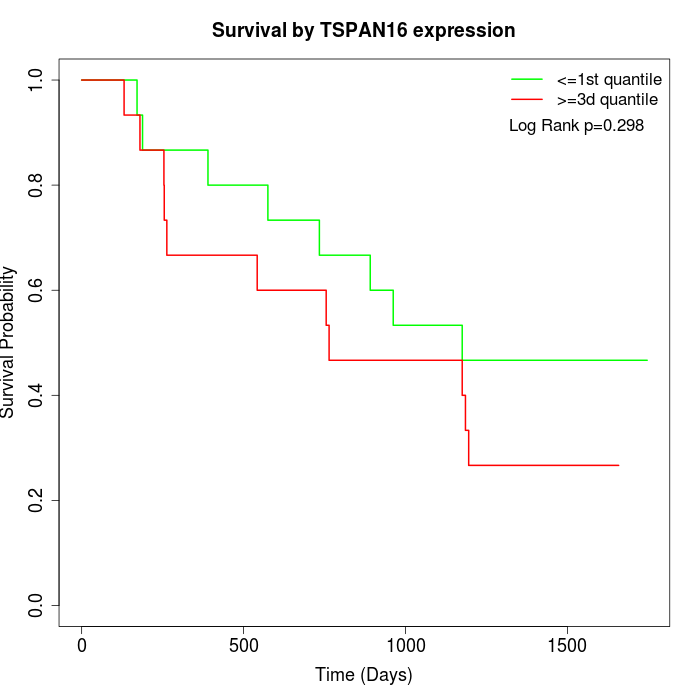
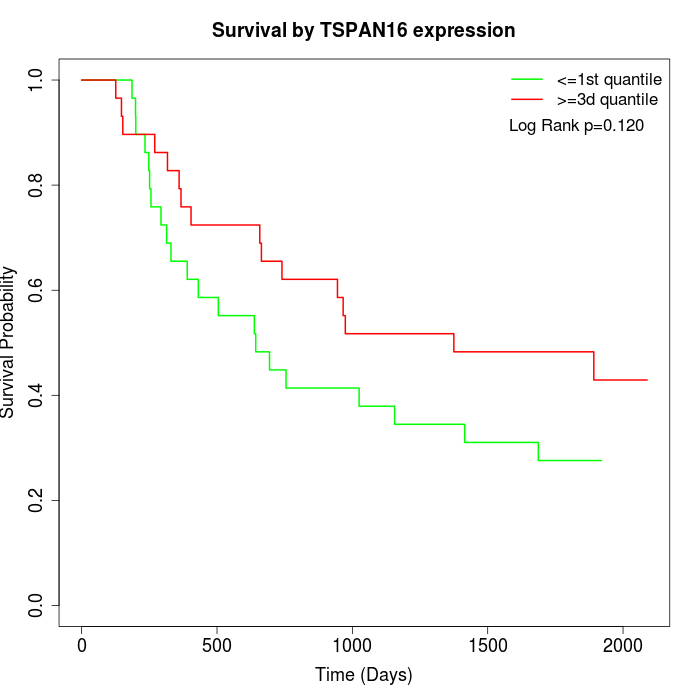
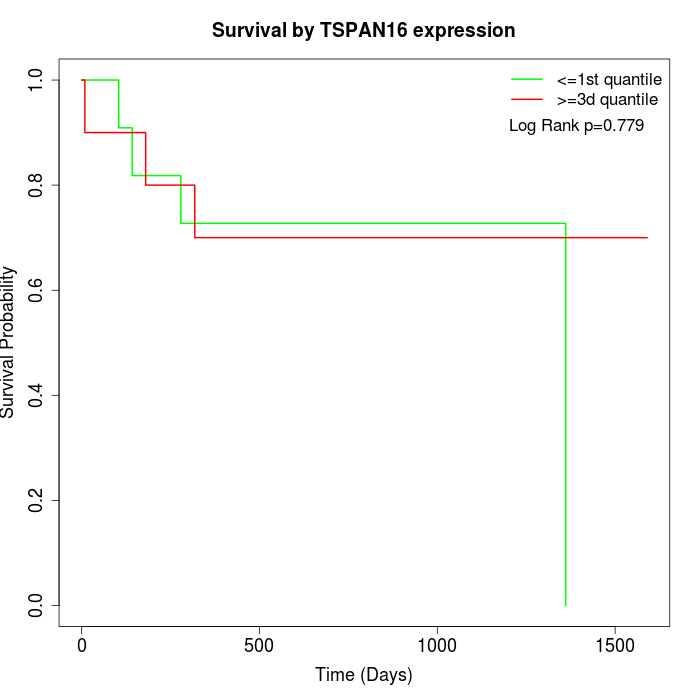
Survival by TSPAN16 expression:
Note: Click image to view full size file.
Copy number change of TSPAN16:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TSPAN16 | 26526 | 4 | 2 | 24 | |
| GSE20123 | TSPAN16 | 26526 | 3 | 1 | 26 | |
| GSE43470 | TSPAN16 | 26526 | 2 | 6 | 35 | |
| GSE46452 | TSPAN16 | 26526 | 47 | 1 | 11 | |
| GSE47630 | TSPAN16 | 26526 | 4 | 7 | 29 | |
| GSE54993 | TSPAN16 | 26526 | 16 | 3 | 51 | |
| GSE54994 | TSPAN16 | 26526 | 6 | 13 | 34 | |
| GSE60625 | TSPAN16 | 26526 | 9 | 0 | 2 | |
| GSE74703 | TSPAN16 | 26526 | 2 | 4 | 30 | |
| GSE74704 | TSPAN16 | 26526 | 0 | 1 | 19 | |
| TCGA | TSPAN16 | 26526 | 17 | 11 | 68 |
Total number of gains: 110; Total number of losses: 49; Total Number of normals: 329.
Somatic mutations of TSPAN16:
Generating mutation plots.
Highly correlated genes for TSPAN16:
Showing top 20/235 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TSPAN16 | C2CD4B | 0.750746 | 3 | 0 | 3 |
| TSPAN16 | JRK | 0.729764 | 3 | 0 | 3 |
| TSPAN16 | IGLL1 | 0.721148 | 3 | 0 | 3 |
| TSPAN16 | KCNQ4 | 0.717783 | 4 | 0 | 4 |
| TSPAN16 | S100A1 | 0.710105 | 3 | 0 | 3 |
| TSPAN16 | XAGE2 | 0.702393 | 3 | 0 | 3 |
| TSPAN16 | SCT | 0.701208 | 3 | 0 | 3 |
| TSPAN16 | GRK1 | 0.696319 | 4 | 0 | 4 |
| TSPAN16 | SLC26A1 | 0.681985 | 5 | 0 | 5 |
| TSPAN16 | CHRNE | 0.678918 | 3 | 0 | 3 |
| TSPAN16 | DPEP3 | 0.67623 | 3 | 0 | 3 |
| TSPAN16 | DNAH9 | 0.672074 | 5 | 0 | 4 |
| TSPAN16 | MYADML2 | 0.670787 | 3 | 0 | 3 |
| TSPAN16 | RASL10B | 0.665004 | 4 | 0 | 4 |
| TSPAN16 | AANAT | 0.662673 | 4 | 0 | 4 |
| TSPAN16 | LIPE-AS1 | 0.659629 | 3 | 0 | 3 |
| TSPAN16 | SHROOM4 | 0.65744 | 3 | 0 | 3 |
| TSPAN16 | WFDC3 | 0.657143 | 3 | 0 | 3 |
| TSPAN16 | PROCA1 | 0.657075 | 5 | 0 | 4 |
| TSPAN16 | TEX26-AS1 | 0.654471 | 3 | 0 | 3 |
For details and further investigation, click here