| Full name: WAP four-disulfide core domain 3 | Alias Symbol: dJ447F3.3|WAP14 | ||
| Type: protein-coding gene | Cytoband: 20q13.12 | ||
| Entrez ID: 140686 | HGNC ID: HGNC:15957 | Ensembl Gene: ENSG00000124116 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of WFDC3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | WFDC3 | 140686 | 232602_at | -0.0392 | 0.9318 | |
| GSE26886 | WFDC3 | 140686 | 232602_at | 0.4811 | 0.0667 | |
| GSE45670 | WFDC3 | 140686 | 232602_at | 0.0683 | 0.7038 | |
| GSE53622 | WFDC3 | 140686 | 145961 | 0.3658 | 0.0000 | |
| GSE53624 | WFDC3 | 140686 | 145961 | 0.5740 | 0.0000 | |
| GSE63941 | WFDC3 | 140686 | 232602_at | -0.3955 | 0.5173 | |
| GSE77861 | WFDC3 | 140686 | 232602_at | -0.0929 | 0.3146 | |
| GSE97050 | WFDC3 | 140686 | A_23_P120435 | 0.0718 | 0.8278 | |
| SRP159526 | WFDC3 | 140686 | RNAseq | 0.2592 | 0.6836 | |
| TCGA | WFDC3 | 140686 | RNAseq | 0.2032 | 0.6945 |
Upregulated datasets: 0; Downregulated datasets: 0.
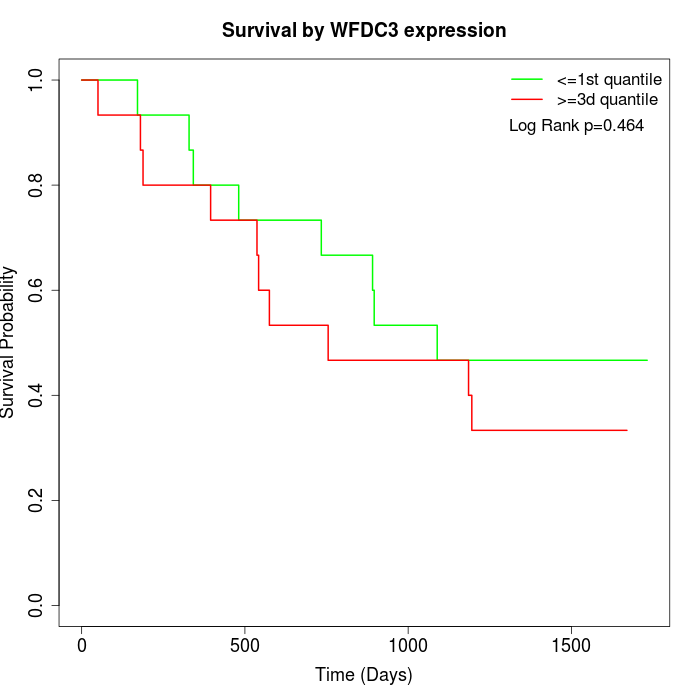
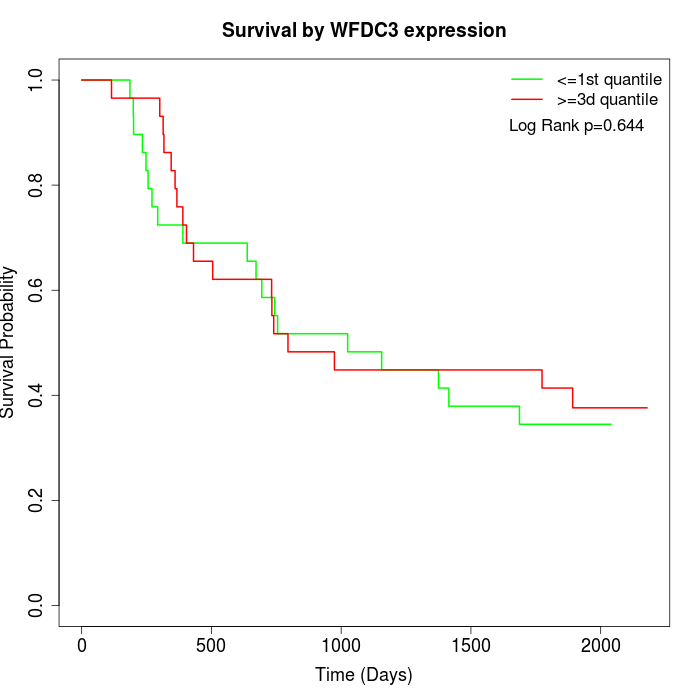
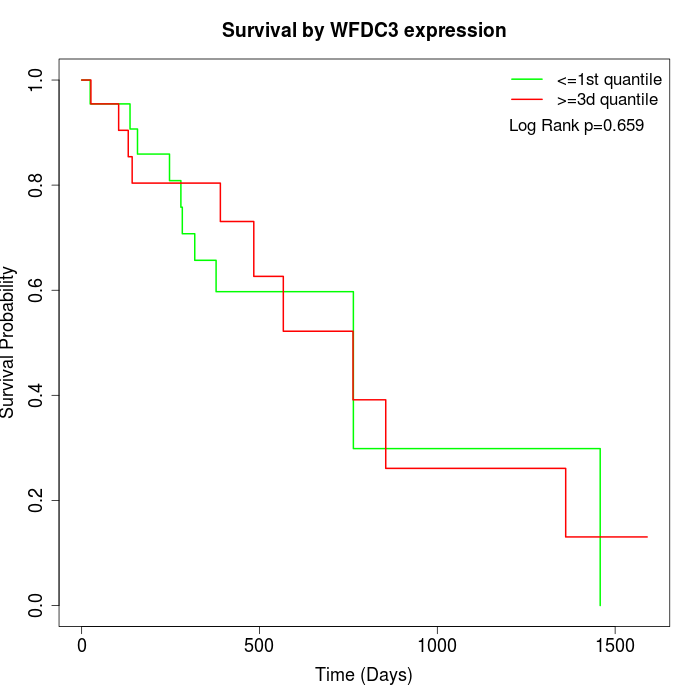
Survival by WFDC3 expression:
Note: Click image to view full size file.
Copy number change of WFDC3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | WFDC3 | 140686 | 14 | 1 | 15 | |
| GSE20123 | WFDC3 | 140686 | 14 | 1 | 15 | |
| GSE43470 | WFDC3 | 140686 | 12 | 1 | 30 | |
| GSE46452 | WFDC3 | 140686 | 29 | 0 | 30 | |
| GSE47630 | WFDC3 | 140686 | 24 | 1 | 15 | |
| GSE54993 | WFDC3 | 140686 | 0 | 18 | 52 | |
| GSE54994 | WFDC3 | 140686 | 25 | 0 | 28 | |
| GSE60625 | WFDC3 | 140686 | 0 | 0 | 11 | |
| GSE74703 | WFDC3 | 140686 | 10 | 1 | 25 | |
| GSE74704 | WFDC3 | 140686 | 10 | 0 | 10 | |
| TCGA | WFDC3 | 140686 | 45 | 3 | 48 |
Total number of gains: 183; Total number of losses: 26; Total Number of normals: 279.
Somatic mutations of WFDC3:
Generating mutation plots.
Highly correlated genes for WFDC3:
Showing top 20/168 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| WFDC3 | TMEM179 | 0.788646 | 3 | 0 | 3 |
| WFDC3 | MGAM | 0.769367 | 3 | 0 | 3 |
| WFDC3 | KCNA10 | 0.764095 | 3 | 0 | 3 |
| WFDC3 | CDKL3 | 0.723603 | 3 | 0 | 3 |
| WFDC3 | BRINP3 | 0.716356 | 4 | 0 | 3 |
| WFDC3 | GABRA5 | 0.713122 | 3 | 0 | 3 |
| WFDC3 | CECR9 | 0.691217 | 4 | 0 | 4 |
| WFDC3 | REM1 | 0.686231 | 3 | 0 | 3 |
| WFDC3 | KCNF1 | 0.685177 | 5 | 0 | 4 |
| WFDC3 | HS6ST3 | 0.678067 | 3 | 0 | 3 |
| WFDC3 | NPY | 0.677616 | 4 | 0 | 3 |
| WFDC3 | ZNF358 | 0.676646 | 3 | 0 | 3 |
| WFDC3 | GDF11 | 0.67258 | 4 | 0 | 4 |
| WFDC3 | F2RL3 | 0.66993 | 4 | 0 | 3 |
| WFDC3 | CDHR5 | 0.663445 | 3 | 0 | 3 |
| WFDC3 | P2RX2 | 0.659614 | 3 | 0 | 3 |
| WFDC3 | GIPC3 | 0.659218 | 4 | 0 | 4 |
| WFDC3 | ZBTB46-AS1 | 0.659107 | 3 | 0 | 3 |
| WFDC3 | TSPAN16 | 0.657143 | 3 | 0 | 3 |
| WFDC3 | ADCY5 | 0.655239 | 3 | 0 | 3 |
For details and further investigation, click here