| Full name: TSR2 ribosome maturation factor | Alias Symbol: DT1P1A10|RP1-112K5.2|WGG1 | ||
| Type: protein-coding gene | Cytoband: Xp11.22 | ||
| Entrez ID: 90121 | HGNC ID: HGNC:25455 | Ensembl Gene: ENSG00000158526 | OMIM ID: 300945 |
| Drug and gene relationship at DGIdb | |||
Expression of TSR2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TSR2 | 90121 | 213079_at | -0.0697 | 0.8864 | |
| GSE20347 | TSR2 | 90121 | 213079_at | 0.0614 | 0.6461 | |
| GSE23400 | TSR2 | 90121 | 213079_at | -0.0286 | 0.6351 | |
| GSE26886 | TSR2 | 90121 | 213079_at | -0.4647 | 0.0139 | |
| GSE29001 | TSR2 | 90121 | 213079_at | -0.2020 | 0.4764 | |
| GSE38129 | TSR2 | 90121 | 213079_at | -0.0352 | 0.7405 | |
| GSE45670 | TSR2 | 90121 | 213079_at | -0.2099 | 0.1756 | |
| GSE53622 | TSR2 | 90121 | 2677 | -0.3413 | 0.0000 | |
| GSE53624 | TSR2 | 90121 | 2677 | -0.1658 | 0.0025 | |
| GSE63941 | TSR2 | 90121 | 213079_at | 0.6542 | 0.0926 | |
| GSE77861 | TSR2 | 90121 | 213079_at | -0.0950 | 0.5888 | |
| GSE97050 | TSR2 | 90121 | A_33_P3341901 | -0.1818 | 0.6098 | |
| SRP007169 | TSR2 | 90121 | RNAseq | 0.3345 | 0.4372 | |
| SRP008496 | TSR2 | 90121 | RNAseq | 0.3545 | 0.2752 | |
| SRP064894 | TSR2 | 90121 | RNAseq | 0.1839 | 0.4128 | |
| SRP133303 | TSR2 | 90121 | RNAseq | 0.2361 | 0.3531 | |
| SRP159526 | TSR2 | 90121 | RNAseq | 0.2769 | 0.2069 | |
| SRP193095 | TSR2 | 90121 | RNAseq | -0.1157 | 0.3021 | |
| SRP219564 | TSR2 | 90121 | RNAseq | -0.0865 | 0.7913 | |
| TCGA | TSR2 | 90121 | RNAseq | -0.0626 | 0.2736 |
Upregulated datasets: 0; Downregulated datasets: 0.
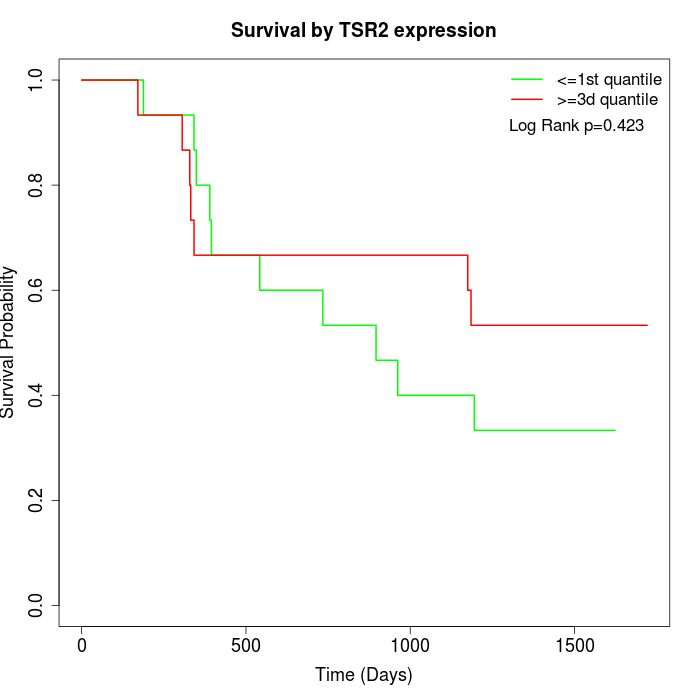
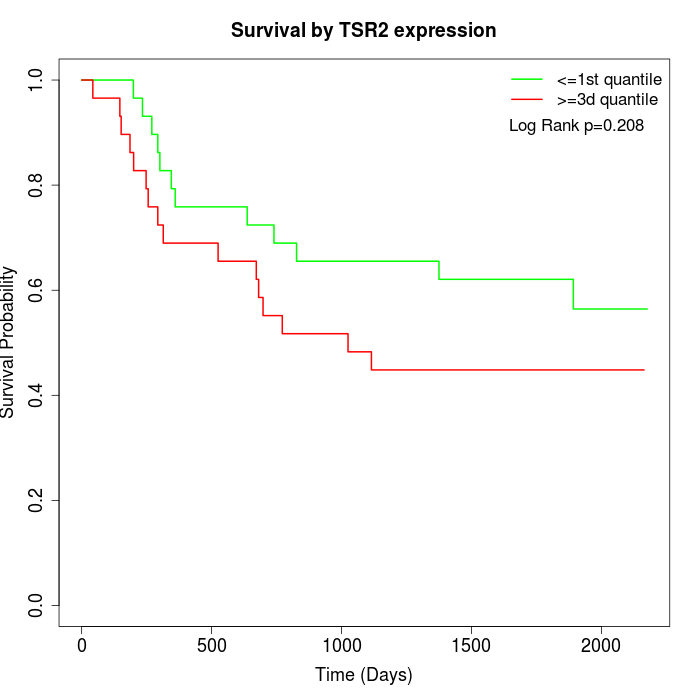
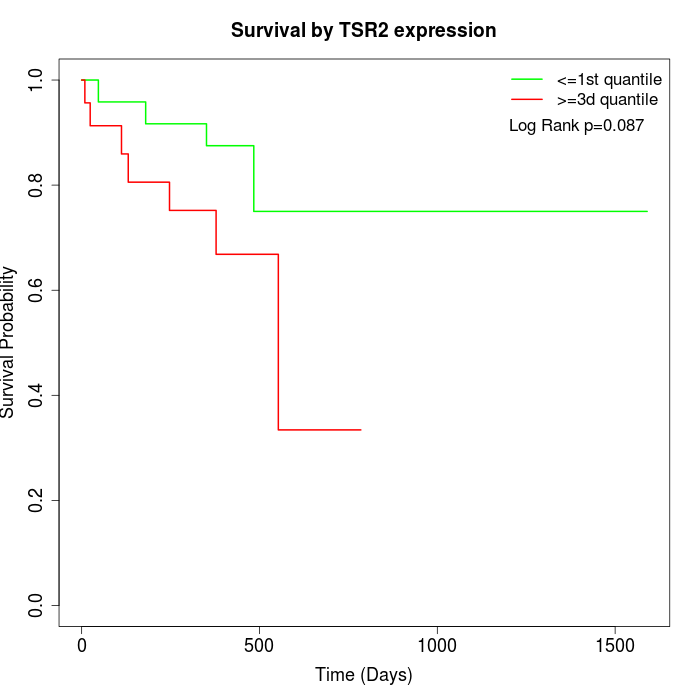
Survival by TSR2 expression:
Note: Click image to view full size file.
Copy number change of TSR2:
No record found for this gene.
Somatic mutations of TSR2:
Generating mutation plots.
Highly correlated genes for TSR2:
Showing top 20/261 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TSR2 | ZIK1 | 0.814387 | 3 | 0 | 3 |
| TSR2 | RNF207 | 0.799646 | 3 | 0 | 3 |
| TSR2 | SPHK2 | 0.776131 | 3 | 0 | 3 |
| TSR2 | EXOC3L1 | 0.765287 | 3 | 0 | 3 |
| TSR2 | KAT5 | 0.762609 | 3 | 0 | 3 |
| TSR2 | ZNF771 | 0.755044 | 3 | 0 | 3 |
| TSR2 | KBTBD4 | 0.750562 | 3 | 0 | 3 |
| TSR2 | HIC2 | 0.749875 | 3 | 0 | 3 |
| TSR2 | CDAN1 | 0.738742 | 3 | 0 | 3 |
| TSR2 | PLIN4 | 0.736819 | 3 | 0 | 3 |
| TSR2 | ZNF214 | 0.724721 | 3 | 0 | 3 |
| TSR2 | ALDH1A1 | 0.723227 | 3 | 0 | 3 |
| TSR2 | TRIM26 | 0.722119 | 3 | 0 | 3 |
| TSR2 | PALM | 0.704588 | 4 | 0 | 3 |
| TSR2 | PDK2 | 0.699274 | 4 | 0 | 3 |
| TSR2 | FAM149A | 0.697828 | 3 | 0 | 3 |
| TSR2 | PITPNM2 | 0.695449 | 3 | 0 | 3 |
| TSR2 | ARHGEF9 | 0.69501 | 3 | 0 | 3 |
| TSR2 | MKRN1 | 0.69334 | 4 | 0 | 3 |
| TSR2 | FBXW8 | 0.691965 | 3 | 0 | 3 |
For details and further investigation, click here