| Full name: lysine acetyltransferase 5 | Alias Symbol: TIP60|PLIP|cPLA2|HTATIP1|ESA1|ZC2HC5 | ||
| Type: protein-coding gene | Cytoband: 11q13.1 | ||
| Entrez ID: 10524 | HGNC ID: HGNC:5275 | Ensembl Gene: ENSG00000172977 | OMIM ID: 601409 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
KAT5 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05166 | HTLV-I infection |
Expression of KAT5:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KAT5 | 10524 | 214258_x_at | -0.3564 | 0.3529 | |
| GSE20347 | KAT5 | 10524 | 214258_x_at | -0.1574 | 0.2126 | |
| GSE23400 | KAT5 | 10524 | 214258_x_at | -0.1607 | 0.0032 | |
| GSE26886 | KAT5 | 10524 | 206689_x_at | -0.0285 | 0.9101 | |
| GSE29001 | KAT5 | 10524 | 214258_x_at | -0.2187 | 0.1411 | |
| GSE38129 | KAT5 | 10524 | 214258_x_at | -0.2469 | 0.0063 | |
| GSE45670 | KAT5 | 10524 | 214258_x_at | -0.1254 | 0.3428 | |
| GSE63941 | KAT5 | 10524 | 214258_x_at | 1.1759 | 0.0113 | |
| GSE77861 | KAT5 | 10524 | 214258_x_at | 0.0190 | 0.8733 | |
| GSE97050 | KAT5 | 10524 | A_23_P138849 | -0.3356 | 0.1623 | |
| SRP007169 | KAT5 | 10524 | RNAseq | -0.7525 | 0.0130 | |
| SRP008496 | KAT5 | 10524 | RNAseq | -0.7304 | 0.0006 | |
| SRP064894 | KAT5 | 10524 | RNAseq | -0.3435 | 0.0043 | |
| SRP133303 | KAT5 | 10524 | RNAseq | -0.1600 | 0.4886 | |
| SRP159526 | KAT5 | 10524 | RNAseq | -0.2908 | 0.0306 | |
| SRP193095 | KAT5 | 10524 | RNAseq | -0.2256 | 0.0066 | |
| SRP219564 | KAT5 | 10524 | RNAseq | -0.4462 | 0.0905 | |
| TCGA | KAT5 | 10524 | RNAseq | -0.0649 | 0.1840 |
Upregulated datasets: 1; Downregulated datasets: 0.
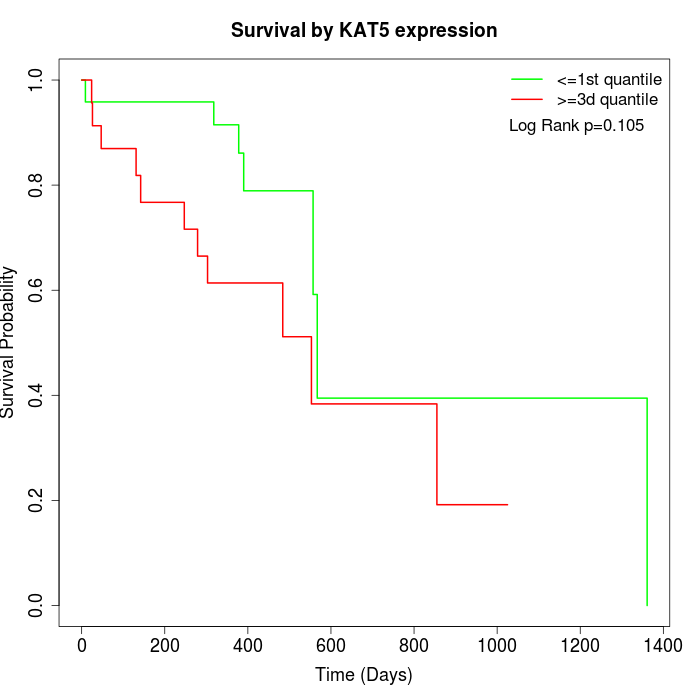
Survival by KAT5 expression:
Note: Click image to view full size file.
Copy number change of KAT5:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KAT5 | 10524 | 8 | 5 | 17 | |
| GSE20123 | KAT5 | 10524 | 8 | 5 | 17 | |
| GSE43470 | KAT5 | 10524 | 6 | 1 | 36 | |
| GSE46452 | KAT5 | 10524 | 11 | 3 | 45 | |
| GSE47630 | KAT5 | 10524 | 7 | 5 | 28 | |
| GSE54993 | KAT5 | 10524 | 3 | 0 | 67 | |
| GSE54994 | KAT5 | 10524 | 7 | 5 | 41 | |
| GSE60625 | KAT5 | 10524 | 0 | 3 | 8 | |
| GSE74703 | KAT5 | 10524 | 4 | 0 | 32 | |
| GSE74704 | KAT5 | 10524 | 6 | 3 | 11 | |
| TCGA | KAT5 | 10524 | 23 | 7 | 66 |
Total number of gains: 83; Total number of losses: 37; Total Number of normals: 368.
Somatic mutations of KAT5:
Generating mutation plots.
Highly correlated genes for KAT5:
Showing top 20/176 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KAT5 | ZBED5 | 0.834617 | 3 | 0 | 3 |
| KAT5 | ALKBH7 | 0.811056 | 3 | 0 | 3 |
| KAT5 | NSMCE4A | 0.806056 | 3 | 0 | 3 |
| KAT5 | ZNF747 | 0.785417 | 3 | 0 | 3 |
| KAT5 | GOT1 | 0.777204 | 3 | 0 | 3 |
| KAT5 | IPO8 | 0.776343 | 3 | 0 | 3 |
| KAT5 | TRIM68 | 0.762741 | 3 | 0 | 3 |
| KAT5 | TSR2 | 0.762609 | 3 | 0 | 3 |
| KAT5 | KCTD15 | 0.757464 | 3 | 0 | 3 |
| KAT5 | FRMD4B | 0.747603 | 3 | 0 | 3 |
| KAT5 | SBF2 | 0.738931 | 3 | 0 | 3 |
| KAT5 | LDB3 | 0.73341 | 3 | 0 | 3 |
| KAT5 | TSHZ1 | 0.733106 | 3 | 0 | 3 |
| KAT5 | SUGP1 | 0.729595 | 3 | 0 | 3 |
| KAT5 | PPP6R3 | 0.728643 | 3 | 0 | 3 |
| KAT5 | THRAP3 | 0.728086 | 3 | 0 | 3 |
| KAT5 | IPO7 | 0.725876 | 3 | 0 | 3 |
| KAT5 | EIF1B | 0.715193 | 3 | 0 | 3 |
| KAT5 | TACC2 | 0.714447 | 4 | 0 | 3 |
| KAT5 | MRPL46 | 0.707241 | 3 | 0 | 3 |
For details and further investigation, click here