| Full name: sphingosine kinase 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 19q13.33 | ||
| Entrez ID: 56848 | HGNC ID: HGNC:18859 | Ensembl Gene: ENSG00000063176 | OMIM ID: 607092 |
| Drug and gene relationship at DGIdb | |||
SPHK2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04020 | Calcium signaling pathway | |
| hsa04071 | Sphingolipid signaling pathway | |
| hsa04370 | VEGF signaling pathway | |
| hsa04666 | Fc gamma R-mediated phagocytosis | |
| hsa05152 | Tuberculosis |
Expression of SPHK2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SPHK2 | 56848 | 40273_at | 0.0451 | 0.9518 | |
| GSE20347 | SPHK2 | 56848 | 40273_at | 0.0509 | 0.5790 | |
| GSE23400 | SPHK2 | 56848 | 209857_s_at | -0.0249 | 0.5656 | |
| GSE26886 | SPHK2 | 56848 | 40273_at | 0.3407 | 0.0218 | |
| GSE29001 | SPHK2 | 56848 | 40273_at | 0.0805 | 0.7662 | |
| GSE38129 | SPHK2 | 56848 | 40273_at | 0.0322 | 0.7374 | |
| GSE45670 | SPHK2 | 56848 | 40273_at | 0.0288 | 0.7975 | |
| GSE53622 | SPHK2 | 56848 | 139190 | 0.0301 | 0.7116 | |
| GSE53624 | SPHK2 | 56848 | 139190 | 0.3422 | 0.0000 | |
| GSE63941 | SPHK2 | 56848 | 209857_s_at | -0.1293 | 0.5850 | |
| GSE77861 | SPHK2 | 56848 | 209857_s_at | -0.0232 | 0.8781 | |
| GSE97050 | SPHK2 | 56848 | A_23_P303181 | 0.1863 | 0.3909 | |
| SRP007169 | SPHK2 | 56848 | RNAseq | 0.0957 | 0.8644 | |
| SRP008496 | SPHK2 | 56848 | RNAseq | -0.1985 | 0.5439 | |
| SRP064894 | SPHK2 | 56848 | RNAseq | 0.3011 | 0.1125 | |
| SRP133303 | SPHK2 | 56848 | RNAseq | -0.2325 | 0.2415 | |
| SRP159526 | SPHK2 | 56848 | RNAseq | 0.2992 | 0.3677 | |
| SRP193095 | SPHK2 | 56848 | RNAseq | -0.1521 | 0.4724 | |
| SRP219564 | SPHK2 | 56848 | RNAseq | 0.2438 | 0.2991 | |
| TCGA | SPHK2 | 56848 | RNAseq | -0.1421 | 0.0149 |
Upregulated datasets: 0; Downregulated datasets: 0.
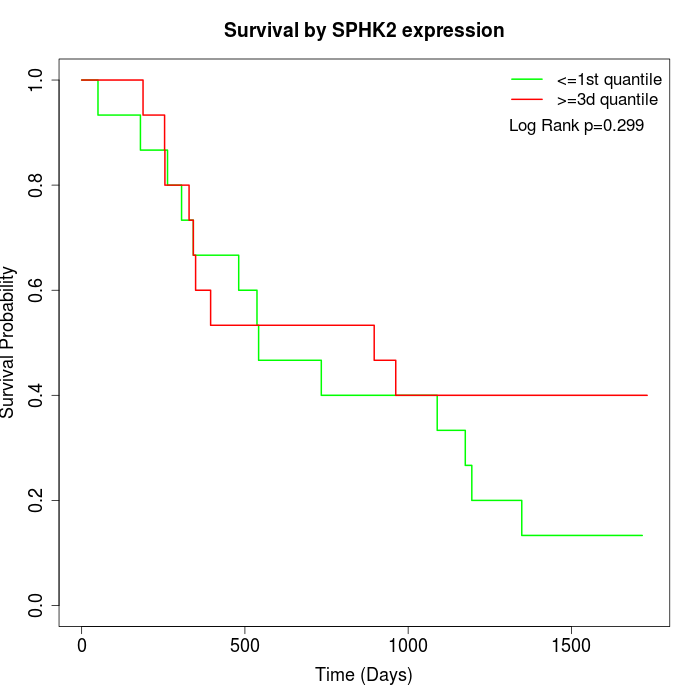
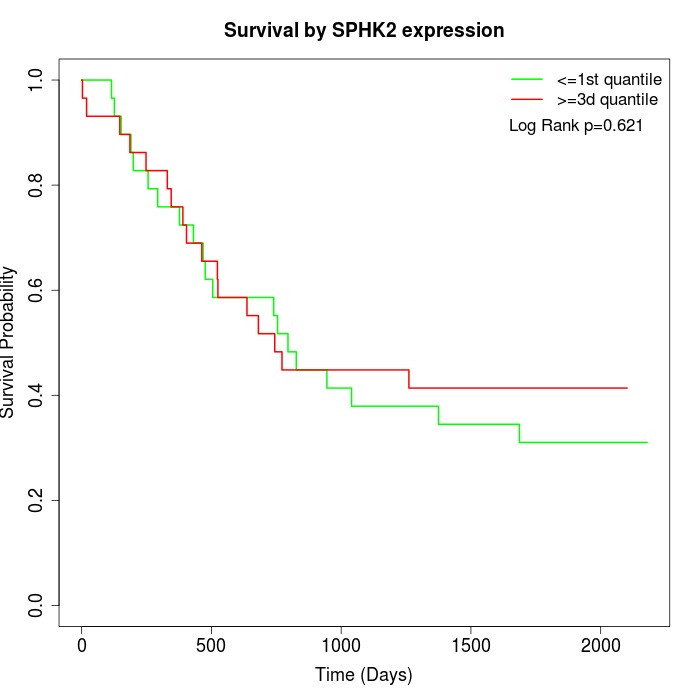
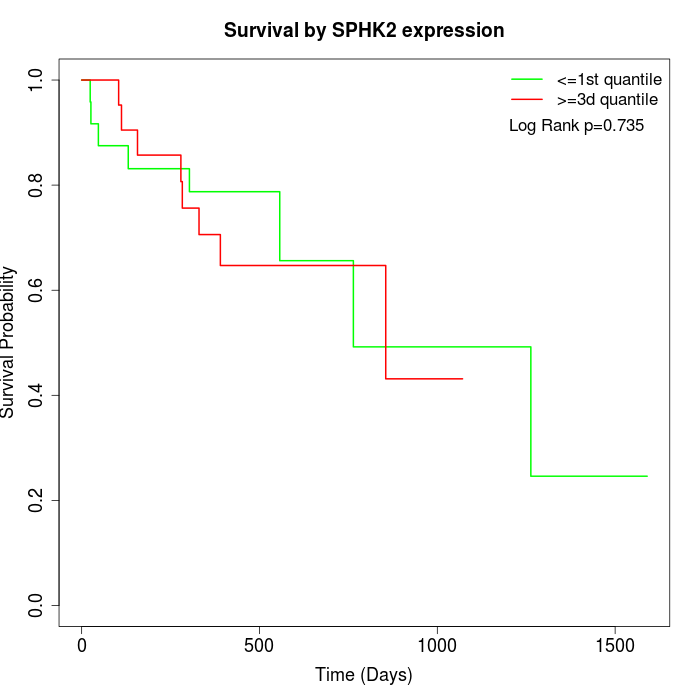
Survival by SPHK2 expression:
Note: Click image to view full size file.
Copy number change of SPHK2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SPHK2 | 56848 | 4 | 4 | 22 | |
| GSE20123 | SPHK2 | 56848 | 4 | 3 | 23 | |
| GSE43470 | SPHK2 | 56848 | 4 | 10 | 29 | |
| GSE46452 | SPHK2 | 56848 | 45 | 1 | 13 | |
| GSE47630 | SPHK2 | 56848 | 8 | 7 | 25 | |
| GSE54993 | SPHK2 | 56848 | 17 | 4 | 49 | |
| GSE54994 | SPHK2 | 56848 | 4 | 14 | 35 | |
| GSE60625 | SPHK2 | 56848 | 9 | 0 | 2 | |
| GSE74703 | SPHK2 | 56848 | 4 | 7 | 25 | |
| GSE74704 | SPHK2 | 56848 | 4 | 1 | 15 | |
| TCGA | SPHK2 | 56848 | 14 | 18 | 64 |
Total number of gains: 117; Total number of losses: 69; Total Number of normals: 302.
Somatic mutations of SPHK2:
Generating mutation plots.
Highly correlated genes for SPHK2:
Showing top 20/370 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SPHK2 | RFNG | 0.777579 | 3 | 0 | 3 |
| SPHK2 | TSR2 | 0.776131 | 3 | 0 | 3 |
| SPHK2 | NDUFB11 | 0.759066 | 3 | 0 | 3 |
| SPHK2 | OS9 | 0.753681 | 3 | 0 | 3 |
| SPHK2 | LRRC41 | 0.753433 | 3 | 0 | 3 |
| SPHK2 | GYG2 | 0.749762 | 3 | 0 | 3 |
| SPHK2 | ZNF837 | 0.748898 | 3 | 0 | 3 |
| SPHK2 | HDAC6 | 0.743929 | 3 | 0 | 3 |
| SPHK2 | NOC2L | 0.73178 | 3 | 0 | 3 |
| SPHK2 | PRKACA | 0.731643 | 4 | 0 | 4 |
| SPHK2 | NR1H2 | 0.728143 | 3 | 0 | 3 |
| SPHK2 | CCDC159 | 0.724049 | 3 | 0 | 3 |
| SPHK2 | ELFN1 | 0.722609 | 3 | 0 | 3 |
| SPHK2 | RAVER1 | 0.72172 | 3 | 0 | 3 |
| SPHK2 | CTXN1 | 0.715414 | 3 | 0 | 3 |
| SPHK2 | TEAD2 | 0.712326 | 4 | 0 | 3 |
| SPHK2 | DENND3 | 0.711977 | 3 | 0 | 3 |
| SPHK2 | THAP7 | 0.702421 | 3 | 0 | 3 |
| SPHK2 | CAMKK2 | 0.701635 | 3 | 0 | 3 |
| SPHK2 | ZNF766 | 0.699119 | 3 | 0 | 3 |
For details and further investigation, click here