| Full name: Cdc42 guanine nucleotide exchange factor 9 | Alias Symbol: KIAA0424|PEM-2 | ||
| Type: protein-coding gene | Cytoband: Xq11.1 | ||
| Entrez ID: 23229 | HGNC ID: HGNC:14561 | Ensembl Gene: ENSG00000131089 | OMIM ID: 300429 |
| Drug and gene relationship at DGIdb | |||
Expression of ARHGEF9:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ARHGEF9 | 23229 | 203264_s_at | -0.4437 | 0.6428 | |
| GSE20347 | ARHGEF9 | 23229 | 203264_s_at | -0.1922 | 0.0459 | |
| GSE23400 | ARHGEF9 | 23229 | 203264_s_at | -0.3869 | 0.0000 | |
| GSE26886 | ARHGEF9 | 23229 | 203263_s_at | -0.0992 | 0.7970 | |
| GSE29001 | ARHGEF9 | 23229 | 203263_s_at | -0.1477 | 0.7604 | |
| GSE38129 | ARHGEF9 | 23229 | 203264_s_at | -0.6317 | 0.0022 | |
| GSE45670 | ARHGEF9 | 23229 | 203264_s_at | -1.3126 | 0.0000 | |
| GSE53622 | ARHGEF9 | 23229 | 42770 | -1.3688 | 0.0000 | |
| GSE53624 | ARHGEF9 | 23229 | 42770 | -1.1141 | 0.0000 | |
| GSE63941 | ARHGEF9 | 23229 | 203264_s_at | -0.0240 | 0.9798 | |
| GSE77861 | ARHGEF9 | 23229 | 203264_s_at | -0.2024 | 0.1269 | |
| GSE97050 | ARHGEF9 | 23229 | A_23_P251705 | -1.1266 | 0.0684 | |
| SRP007169 | ARHGEF9 | 23229 | RNAseq | -0.3539 | 0.5003 | |
| SRP008496 | ARHGEF9 | 23229 | RNAseq | -0.0174 | 0.9625 | |
| SRP064894 | ARHGEF9 | 23229 | RNAseq | -0.3652 | 0.1812 | |
| SRP133303 | ARHGEF9 | 23229 | RNAseq | -0.3842 | 0.1781 | |
| SRP159526 | ARHGEF9 | 23229 | RNAseq | -0.5677 | 0.2516 | |
| SRP193095 | ARHGEF9 | 23229 | RNAseq | -0.2827 | 0.0607 | |
| SRP219564 | ARHGEF9 | 23229 | RNAseq | -1.0371 | 0.0270 | |
| TCGA | ARHGEF9 | 23229 | RNAseq | -0.5495 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 4.
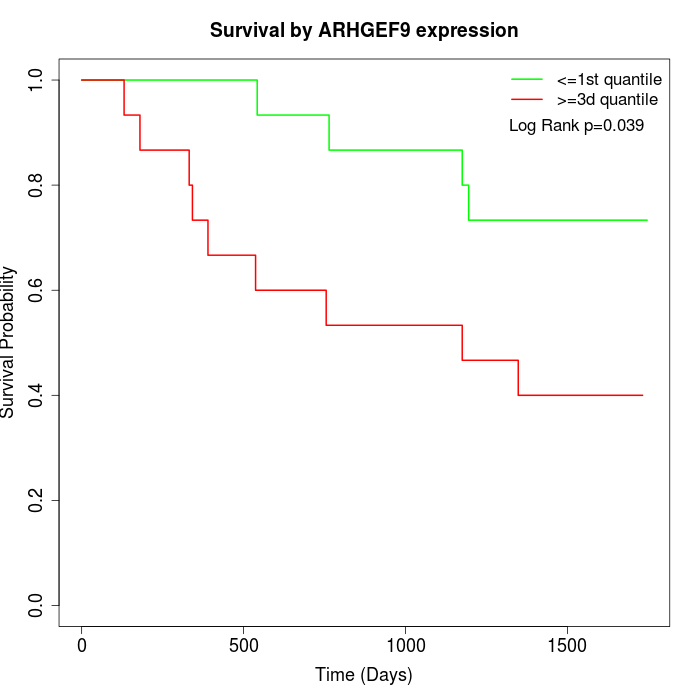
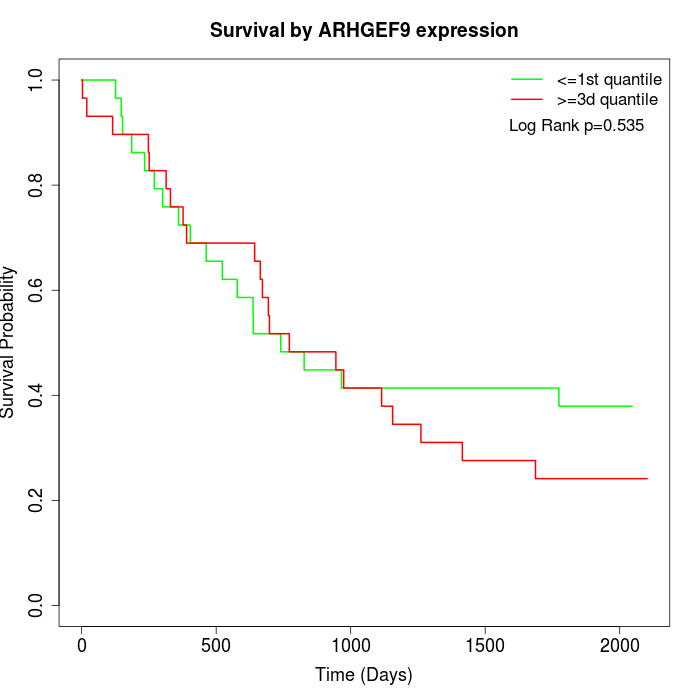
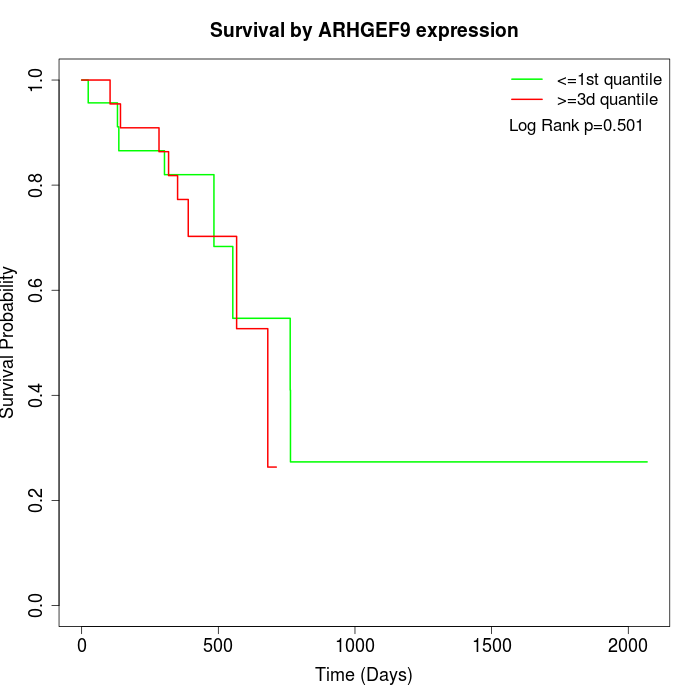
Survival by ARHGEF9 expression:
Note: Click image to view full size file.
Copy number change of ARHGEF9:
No record found for this gene.
Somatic mutations of ARHGEF9:
Generating mutation plots.
Highly correlated genes for ARHGEF9:
Showing top 20/1191 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ARHGEF9 | CYP4V2 | 0.791708 | 3 | 0 | 3 |
| ARHGEF9 | PARD3B | 0.78785 | 4 | 0 | 4 |
| ARHGEF9 | FHL1 | 0.780016 | 7 | 0 | 7 |
| ARHGEF9 | MRVI1 | 0.777433 | 5 | 0 | 5 |
| ARHGEF9 | ANGPTL1 | 0.774733 | 5 | 0 | 5 |
| ARHGEF9 | SYNPO2 | 0.771072 | 5 | 0 | 5 |
| ARHGEF9 | CYP21A2 | 0.762792 | 3 | 0 | 3 |
| ARHGEF9 | NEGR1 | 0.759312 | 5 | 0 | 5 |
| ARHGEF9 | SELENBP1 | 0.758457 | 9 | 0 | 9 |
| ARHGEF9 | C1QTNF7 | 0.757581 | 5 | 0 | 5 |
| ARHGEF9 | BHMT2 | 0.755321 | 7 | 0 | 7 |
| ARHGEF9 | CLEC3B | 0.755281 | 3 | 0 | 3 |
| ARHGEF9 | ING3 | 0.750598 | 3 | 0 | 3 |
| ARHGEF9 | AFF3 | 0.748775 | 3 | 0 | 3 |
| ARHGEF9 | SPARCL1 | 0.748602 | 7 | 0 | 7 |
| ARHGEF9 | RNF150 | 0.743548 | 5 | 0 | 5 |
| ARHGEF9 | RBPMS2 | 0.740793 | 5 | 0 | 5 |
| ARHGEF9 | GPRASP2 | 0.738759 | 3 | 0 | 3 |
| ARHGEF9 | PEG3 | 0.737474 | 5 | 0 | 5 |
| ARHGEF9 | CBX7 | 0.735125 | 9 | 0 | 8 |
For details and further investigation, click here