| Full name: testis-specific transcript, Y-linked 12 | Alias Symbol: TTY11|NCRNA00135 | ||
| Type: non-coding RNA | Cytoband: Yp11.2 | ||
| Entrez ID: 83867 | HGNC ID: HGNC:18493 | Ensembl Gene: ENSG00000237048 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of TTTY12:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TTTY12 | 83867 | 224195_at | -0.0420 | 0.8497 | |
| GSE26886 | TTTY12 | 83867 | 224195_at | -0.0903 | 0.2705 | |
| GSE45670 | TTTY12 | 83867 | 224195_at | 0.0209 | 0.8249 | |
| GSE53622 | TTTY12 | 83867 | 71603 | 0.1246 | 0.2319 | |
| GSE53624 | TTTY12 | 83867 | 71603 | 0.0243 | 0.8695 | |
| GSE63941 | TTTY12 | 83867 | 224195_at | 0.0431 | 0.7169 | |
| GSE77861 | TTTY12 | 83867 | 224195_at | -0.1358 | 0.1101 |
Upregulated datasets: 0; Downregulated datasets: 0.
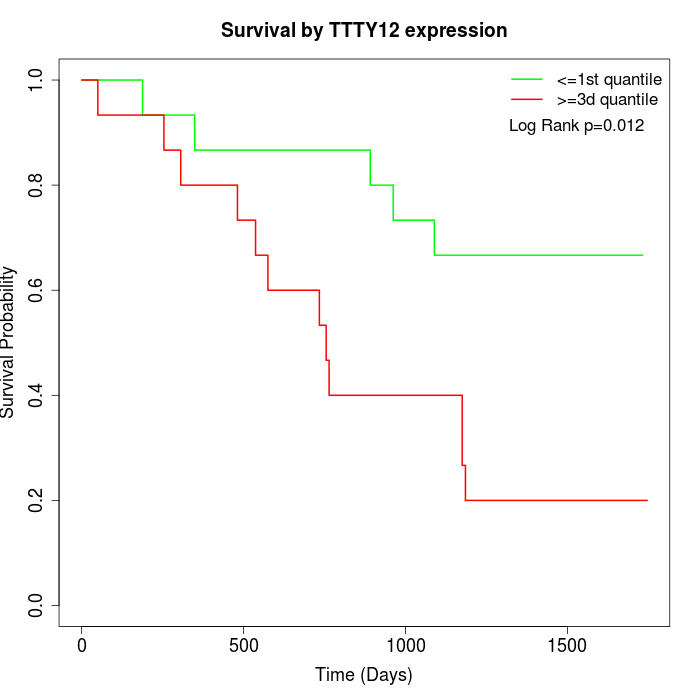
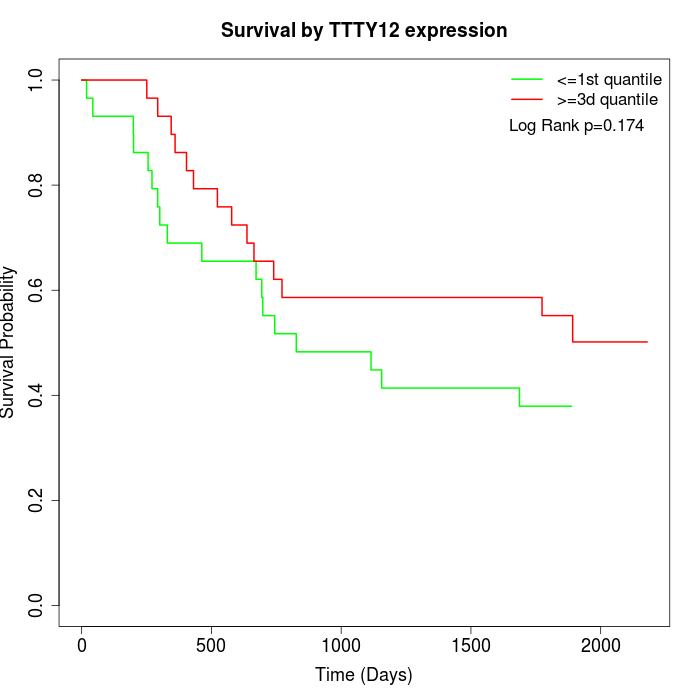
Survival by TTTY12 expression:
Note: Click image to view full size file.
Copy number change of TTTY12:
No record found for this gene.
Somatic mutations of TTTY12:
Generating mutation plots.
Highly correlated genes for TTTY12:
Showing top 20/45 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TTTY12 | ZNF619 | 0.652211 | 3 | 0 | 3 |
| TTTY12 | DEFB121 | 0.650599 | 4 | 0 | 3 |
| TTTY12 | TEX28 | 0.625856 | 3 | 0 | 3 |
| TTTY12 | OR51M1 | 0.625569 | 4 | 0 | 3 |
| TTTY12 | ASB16 | 0.619331 | 4 | 0 | 3 |
| TTTY12 | CRYBB1 | 0.619136 | 3 | 0 | 3 |
| TTTY12 | BTNL2 | 0.618952 | 5 | 0 | 4 |
| TTTY12 | LINC00620 | 0.614717 | 4 | 0 | 3 |
| TTTY12 | GPR15 | 0.600593 | 3 | 0 | 3 |
| TTTY12 | C20orf85 | 0.596833 | 4 | 0 | 3 |
| TTTY12 | ST8SIA3 | 0.594216 | 4 | 0 | 3 |
| TTTY12 | RFFL | 0.592688 | 3 | 0 | 3 |
| TTTY12 | SH3GL3 | 0.589512 | 3 | 0 | 3 |
| TTTY12 | CAMK2A | 0.588408 | 3 | 0 | 3 |
| TTTY12 | LINC00705 | 0.587934 | 4 | 0 | 3 |
| TTTY12 | WFDC9 | 0.579749 | 4 | 0 | 3 |
| TTTY12 | C2orf72 | 0.57864 | 4 | 0 | 4 |
| TTTY12 | CNR2 | 0.576696 | 4 | 0 | 3 |
| TTTY12 | TBATA | 0.576632 | 3 | 0 | 3 |
| TTTY12 | MALRD1 | 0.576024 | 3 | 0 | 3 |
For details and further investigation, click here