| Full name: cannabinoid receptor 2 | Alias Symbol: CB2 | ||
| Type: protein-coding gene | Cytoband: 1p36.11 | ||
| Entrez ID: 1269 | HGNC ID: HGNC:2160 | Ensembl Gene: ENSG00000188822 | OMIM ID: 605051 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of CNR2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CNR2 | 1269 | 206586_at | -0.3184 | 0.3490 | |
| GSE20347 | CNR2 | 1269 | 206586_at | -0.1133 | 0.3190 | |
| GSE23400 | CNR2 | 1269 | 206586_at | -0.1750 | 0.0001 | |
| GSE26886 | CNR2 | 1269 | 206586_at | -0.5329 | 0.0017 | |
| GSE29001 | CNR2 | 1269 | 206586_at | -0.2793 | 0.2732 | |
| GSE38129 | CNR2 | 1269 | 206586_at | -0.2137 | 0.0074 | |
| GSE45670 | CNR2 | 1269 | 206586_at | 0.0313 | 0.8329 | |
| GSE53622 | CNR2 | 1269 | 37369 | -0.7585 | 0.0000 | |
| GSE53624 | CNR2 | 1269 | 37369 | -0.7627 | 0.0000 | |
| GSE63941 | CNR2 | 1269 | 206586_at | 0.4372 | 0.0409 | |
| GSE77861 | CNR2 | 1269 | 206586_at | -0.2309 | 0.0253 | |
| GSE97050 | CNR2 | 1269 | A_23_P310931 | 0.5297 | 0.5355 | |
| SRP007169 | CNR2 | 1269 | RNAseq | -0.4295 | 0.6617 | |
| SRP133303 | CNR2 | 1269 | RNAseq | -0.3564 | 0.0612 | |
| SRP159526 | CNR2 | 1269 | RNAseq | -1.6991 | 0.0224 | |
| SRP193095 | CNR2 | 1269 | RNAseq | -0.5163 | 0.0147 | |
| SRP219564 | CNR2 | 1269 | RNAseq | -0.3165 | 0.5248 | |
| TCGA | CNR2 | 1269 | RNAseq | -1.2143 | 0.1434 |
Upregulated datasets: 0; Downregulated datasets: 1.
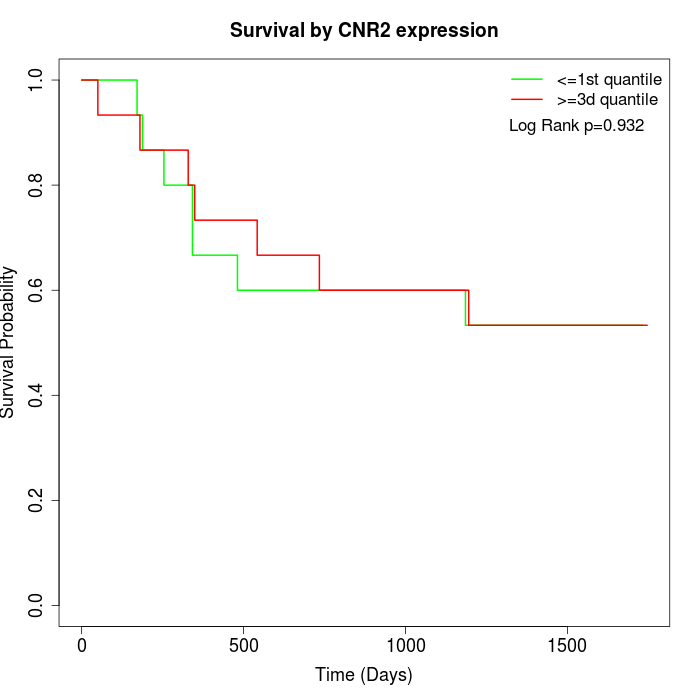
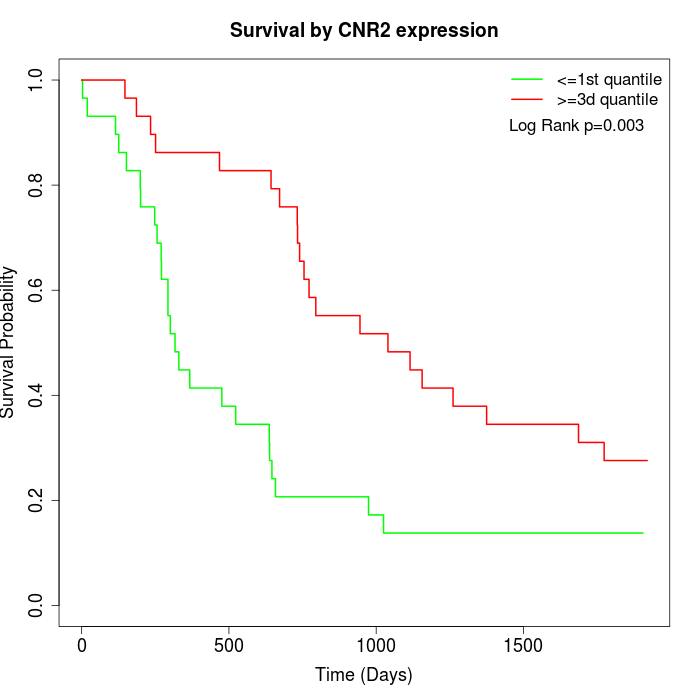
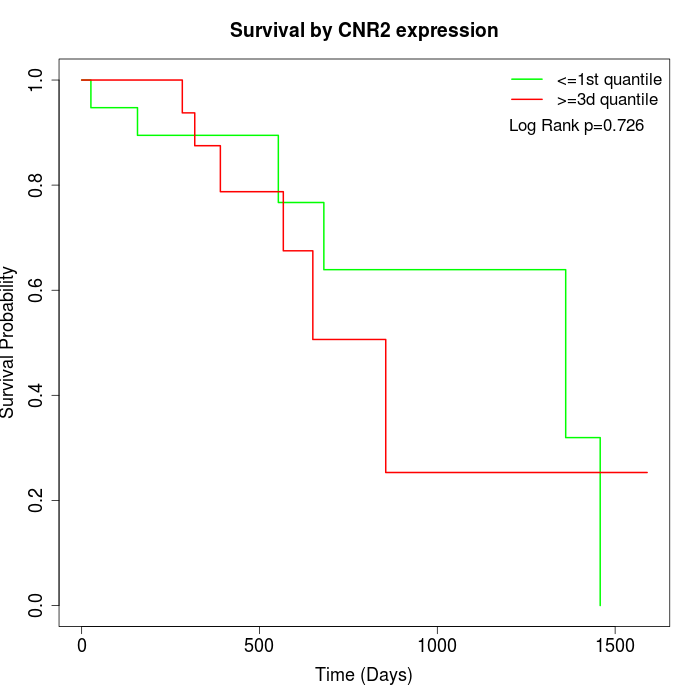
Survival by CNR2 expression:
Note: Click image to view full size file.
Copy number change of CNR2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CNR2 | 1269 | 0 | 5 | 25 | |
| GSE20123 | CNR2 | 1269 | 0 | 4 | 26 | |
| GSE43470 | CNR2 | 1269 | 2 | 6 | 35 | |
| GSE46452 | CNR2 | 1269 | 5 | 1 | 53 | |
| GSE47630 | CNR2 | 1269 | 8 | 3 | 29 | |
| GSE54993 | CNR2 | 1269 | 2 | 1 | 67 | |
| GSE54994 | CNR2 | 1269 | 11 | 4 | 38 | |
| GSE60625 | CNR2 | 1269 | 0 | 0 | 11 | |
| GSE74703 | CNR2 | 1269 | 1 | 4 | 31 | |
| GSE74704 | CNR2 | 1269 | 0 | 0 | 20 | |
| TCGA | CNR2 | 1269 | 9 | 24 | 63 |
Total number of gains: 38; Total number of losses: 52; Total Number of normals: 398.
Somatic mutations of CNR2:
Generating mutation plots.
Highly correlated genes for CNR2:
Showing top 20/1121 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CNR2 | MEPE | 0.720664 | 4 | 0 | 4 |
| CNR2 | GABRR2 | 0.715784 | 4 | 0 | 4 |
| CNR2 | GDF3 | 0.709318 | 5 | 0 | 5 |
| CNR2 | ITGA10 | 0.707911 | 6 | 0 | 6 |
| CNR2 | EPS8L3 | 0.704913 | 6 | 0 | 6 |
| CNR2 | RORC | 0.704202 | 9 | 0 | 9 |
| CNR2 | PSG7 | 0.703742 | 5 | 0 | 4 |
| CNR2 | SRRM2-AS1 | 0.700041 | 3 | 0 | 3 |
| CNR2 | OLFM1 | 0.699348 | 3 | 0 | 3 |
| CNR2 | CACNG6 | 0.697809 | 5 | 0 | 5 |
| CNR2 | PF4 | 0.696234 | 6 | 0 | 6 |
| CNR2 | CALCR | 0.694285 | 6 | 0 | 6 |
| CNR2 | ZNF548 | 0.694139 | 3 | 0 | 3 |
| CNR2 | EDA2R | 0.690541 | 7 | 0 | 6 |
| CNR2 | NR1H4 | 0.687581 | 4 | 0 | 4 |
| CNR2 | ASIC2 | 0.6867 | 5 | 0 | 4 |
| CNR2 | CYP4A11 | 0.685971 | 8 | 0 | 8 |
| CNR2 | AKAP6 | 0.685553 | 5 | 0 | 5 |
| CNR2 | CYP2A7 | 0.685479 | 7 | 0 | 7 |
| CNR2 | OR2H2 | 0.684762 | 5 | 0 | 5 |
For details and further investigation, click here