| Full name: terminal uridylyl transferase 1, U6 snRNA-specific | Alias Symbol: FLJ22347|FLJ22267|FLJ21850|PAPD2|TUTase|TENT1 | ||
| Type: protein-coding gene | Cytoband: 11q12.3 | ||
| Entrez ID: 64852 | HGNC ID: HGNC:26184 | Ensembl Gene: ENSG00000149016 | OMIM ID: 610641 |
| Drug and gene relationship at DGIdb | |||
Expression of TUT1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TUT1 | 64852 | 218965_s_at | 0.0413 | 0.8877 | |
| GSE20347 | TUT1 | 64852 | 218965_s_at | 0.0502 | 0.5744 | |
| GSE23400 | TUT1 | 64852 | 218965_s_at | -0.0039 | 0.9317 | |
| GSE26886 | TUT1 | 64852 | 218965_s_at | 0.4407 | 0.0003 | |
| GSE29001 | TUT1 | 64852 | 218965_s_at | -0.0094 | 0.9595 | |
| GSE38129 | TUT1 | 64852 | 218965_s_at | -0.0454 | 0.5104 | |
| GSE45670 | TUT1 | 64852 | 218965_s_at | 0.2031 | 0.0021 | |
| GSE53622 | TUT1 | 64852 | 72371 | 0.2983 | 0.0000 | |
| GSE53624 | TUT1 | 64852 | 72371 | 0.4321 | 0.0000 | |
| GSE63941 | TUT1 | 64852 | 218965_s_at | 0.2750 | 0.1530 | |
| GSE77861 | TUT1 | 64852 | 218965_s_at | -0.1062 | 0.6408 | |
| GSE97050 | TUT1 | 64852 | A_23_P53015 | -0.0632 | 0.7575 | |
| SRP007169 | TUT1 | 64852 | RNAseq | -0.2750 | 0.5096 | |
| SRP008496 | TUT1 | 64852 | RNAseq | -0.1148 | 0.7271 | |
| SRP064894 | TUT1 | 64852 | RNAseq | 0.3581 | 0.0084 | |
| SRP133303 | TUT1 | 64852 | RNAseq | 0.0214 | 0.8952 | |
| SRP159526 | TUT1 | 64852 | RNAseq | 0.4866 | 0.0605 | |
| SRP193095 | TUT1 | 64852 | RNAseq | 0.3730 | 0.0006 | |
| SRP219564 | TUT1 | 64852 | RNAseq | 0.3011 | 0.3318 | |
| TCGA | TUT1 | 64852 | RNAseq | -0.0253 | 0.6512 |
Upregulated datasets: 0; Downregulated datasets: 0.
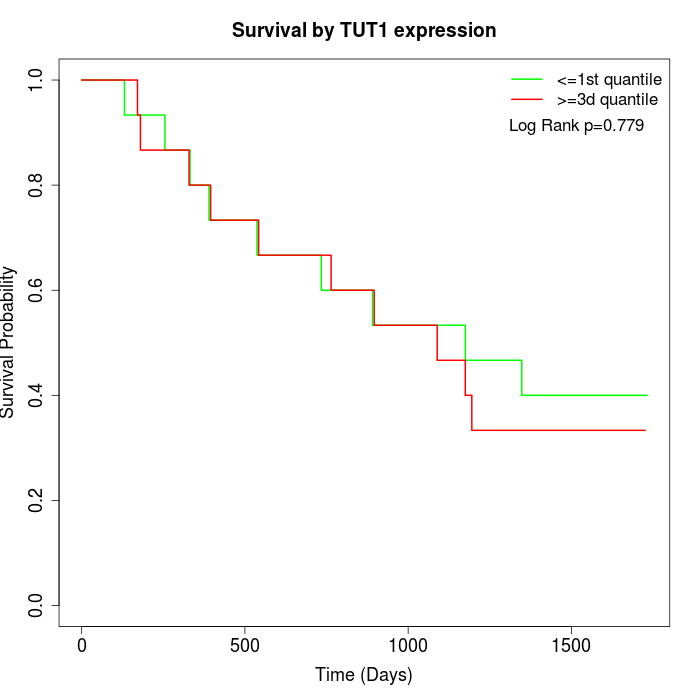
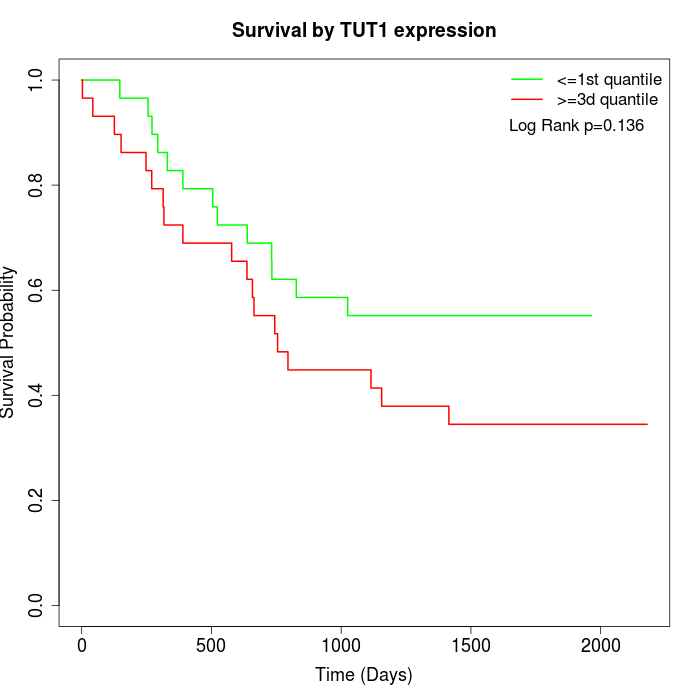
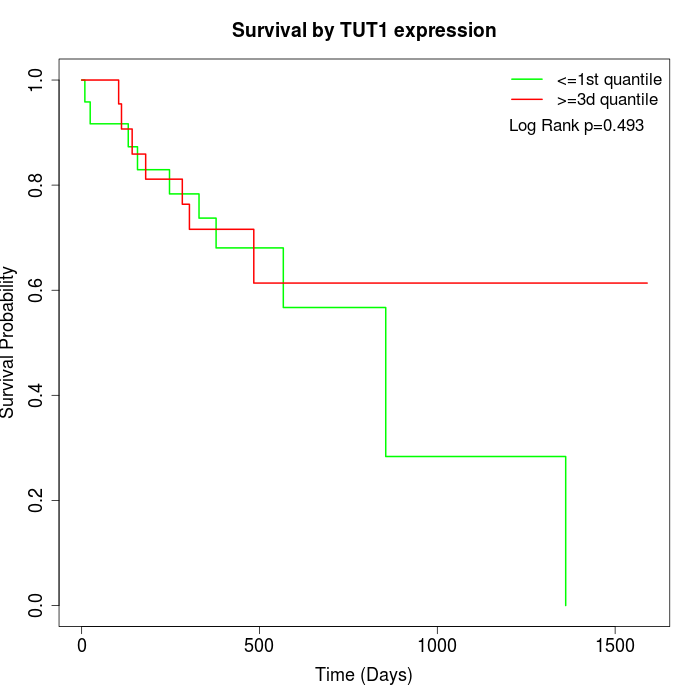
Survival by TUT1 expression:
Note: Click image to view full size file.
Copy number change of TUT1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TUT1 | 64852 | 6 | 5 | 19 | |
| GSE20123 | TUT1 | 64852 | 6 | 5 | 19 | |
| GSE43470 | TUT1 | 64852 | 1 | 6 | 36 | |
| GSE46452 | TUT1 | 64852 | 8 | 4 | 47 | |
| GSE47630 | TUT1 | 64852 | 3 | 7 | 30 | |
| GSE54993 | TUT1 | 64852 | 3 | 0 | 67 | |
| GSE54994 | TUT1 | 64852 | 4 | 5 | 44 | |
| GSE60625 | TUT1 | 64852 | 0 | 3 | 8 | |
| GSE74703 | TUT1 | 64852 | 1 | 3 | 32 | |
| GSE74704 | TUT1 | 64852 | 4 | 3 | 13 | |
| TCGA | TUT1 | 64852 | 14 | 11 | 71 |
Total number of gains: 50; Total number of losses: 52; Total Number of normals: 386.
Somatic mutations of TUT1:
Generating mutation plots.
Highly correlated genes for TUT1:
Showing top 20/496 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TUT1 | JSRP1 | 0.829216 | 3 | 0 | 3 |
| TUT1 | BSND | 0.81891 | 3 | 0 | 3 |
| TUT1 | ZNF219 | 0.808832 | 3 | 0 | 3 |
| TUT1 | DUSP15 | 0.797385 | 3 | 0 | 3 |
| TUT1 | C19orf44 | 0.787317 | 3 | 0 | 3 |
| TUT1 | KLK2 | 0.781943 | 3 | 0 | 3 |
| TUT1 | SFTPC | 0.771105 | 3 | 0 | 3 |
| TUT1 | MGAT5B | 0.765586 | 3 | 0 | 3 |
| TUT1 | REG3A | 0.753355 | 4 | 0 | 4 |
| TUT1 | SPACA3 | 0.752359 | 3 | 0 | 3 |
| TUT1 | STK32C | 0.751884 | 4 | 0 | 4 |
| TUT1 | PPP1R3F | 0.740902 | 3 | 0 | 3 |
| TUT1 | LRRC46 | 0.73751 | 3 | 0 | 3 |
| TUT1 | NKAIN4 | 0.734854 | 5 | 0 | 4 |
| TUT1 | KIRREL2 | 0.729823 | 3 | 0 | 3 |
| TUT1 | SULT1C3 | 0.72767 | 3 | 0 | 3 |
| TUT1 | OSBPL5 | 0.72699 | 4 | 0 | 3 |
| TUT1 | GRAP | 0.723131 | 3 | 0 | 3 |
| TUT1 | COL24A1 | 0.715874 | 3 | 0 | 3 |
| TUT1 | ACSF3 | 0.709592 | 3 | 0 | 3 |
For details and further investigation, click here