| Full name: X antigen family member 3 | Alias Symbol: XAGE-3|pp9012|CT12.3a|CT12.3b | ||
| Type: protein-coding gene | Cytoband: Xp11.22 | ||
| Entrez ID: 170626 | HGNC ID: HGNC:14618 | Ensembl Gene: ENSG00000171402 | OMIM ID: 300740 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of XAGE3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | XAGE3 | 170626 | 236040_at | 0.1215 | 0.5592 | |
| GSE26886 | XAGE3 | 170626 | 236040_at | -0.0154 | 0.9132 | |
| GSE45670 | XAGE3 | 170626 | 236040_at | 0.1095 | 0.3281 | |
| GSE53622 | XAGE3 | 170626 | 38034 | 0.2412 | 0.0926 | |
| GSE53624 | XAGE3 | 170626 | 38034 | 0.1953 | 0.0753 | |
| GSE63941 | XAGE3 | 170626 | 236040_at | -0.0142 | 0.9464 | |
| GSE77861 | XAGE3 | 170626 | 236040_at | -0.2483 | 0.0846 |
Upregulated datasets: 0; Downregulated datasets: 0.
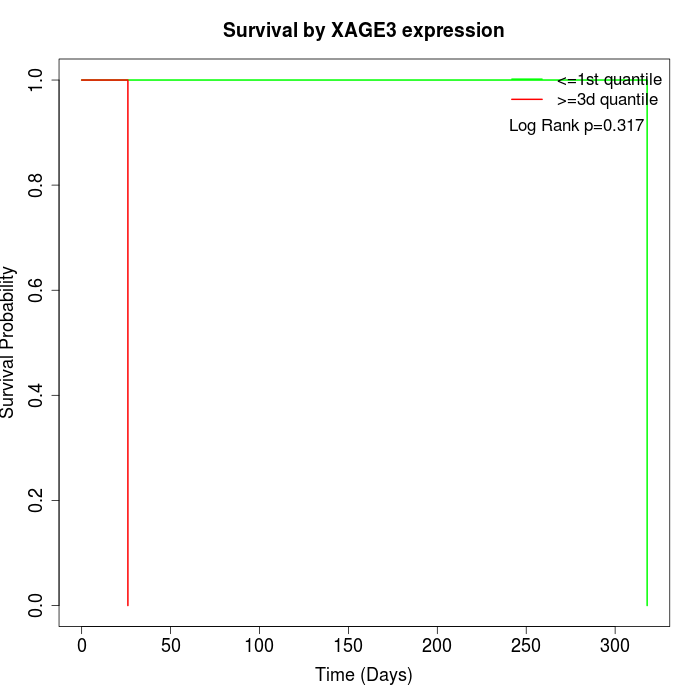
Survival by XAGE3 expression:
Note: Click image to view full size file.
Copy number change of XAGE3:
No record found for this gene.
Somatic mutations of XAGE3:
Generating mutation plots.
Highly correlated genes for XAGE3:
Showing top 20/21 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| XAGE3 | SPATA3 | 0.656902 | 3 | 0 | 3 |
| XAGE3 | CDK5R2 | 0.60758 | 3 | 0 | 3 |
| XAGE3 | RS1 | 0.595978 | 4 | 0 | 3 |
| XAGE3 | UBTD1 | 0.595062 | 3 | 0 | 3 |
| XAGE3 | BFSP2-AS1 | 0.585203 | 3 | 0 | 3 |
| XAGE3 | ANKUB1 | 0.571483 | 3 | 0 | 3 |
| XAGE3 | C21orf58 | 0.57072 | 3 | 0 | 3 |
| XAGE3 | OTOP2 | 0.565431 | 4 | 0 | 3 |
| XAGE3 | ZP2 | 0.564225 | 4 | 0 | 3 |
| XAGE3 | FCAR | 0.56078 | 3 | 0 | 3 |
| XAGE3 | C7orf61 | 0.555868 | 3 | 0 | 3 |
| XAGE3 | LILRB4 | 0.55049 | 3 | 0 | 3 |
| XAGE3 | MBD3L1 | 0.53908 | 4 | 0 | 3 |
| XAGE3 | OPRL1 | 0.536941 | 4 | 0 | 3 |
| XAGE3 | CLECL1 | 0.53593 | 5 | 0 | 3 |
| XAGE3 | ZGLP1 | 0.534174 | 4 | 0 | 3 |
| XAGE3 | SLC26A1 | 0.533083 | 4 | 0 | 3 |
| XAGE3 | ST8SIA3 | 0.527754 | 4 | 0 | 3 |
| XAGE3 | CBLN4 | 0.52625 | 5 | 0 | 3 |
| XAGE3 | WFDC8 | 0.511027 | 5 | 0 | 3 |
For details and further investigation, click here