| Full name: C-type lectin like 1 | Alias Symbol: DCAL1 | ||
| Type: protein-coding gene | Cytoband: 12p13.31 | ||
| Entrez ID: 160365 | HGNC ID: HGNC:24462 | Ensembl Gene: ENSG00000184293 | OMIM ID: 607467 |
| Drug and gene relationship at DGIdb | |||
Expression of CLECL1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CLECL1 | 160365 | 244413_at | 0.3361 | 0.7753 | |
| GSE26886 | CLECL1 | 160365 | 1561899_at | -0.2175 | 0.2351 | |
| GSE45670 | CLECL1 | 160365 | 244413_at | 0.0553 | 0.7437 | |
| GSE53622 | CLECL1 | 160365 | 113751 | 0.0658 | 0.4005 | |
| GSE53624 | CLECL1 | 160365 | 113751 | -0.1133 | 0.2013 | |
| GSE63941 | CLECL1 | 160365 | 244413_at | -0.1105 | 0.3153 | |
| GSE77861 | CLECL1 | 160365 | 1561899_at | -0.0384 | 0.8521 | |
| GSE97050 | CLECL1 | 160365 | A_23_P321984 | 0.2673 | 0.3739 | |
| SRP133303 | CLECL1 | 160365 | RNAseq | -0.5338 | 0.1314 | |
| TCGA | CLECL1 | 160365 | RNAseq | 0.5156 | 0.2475 |
Upregulated datasets: 0; Downregulated datasets: 0.
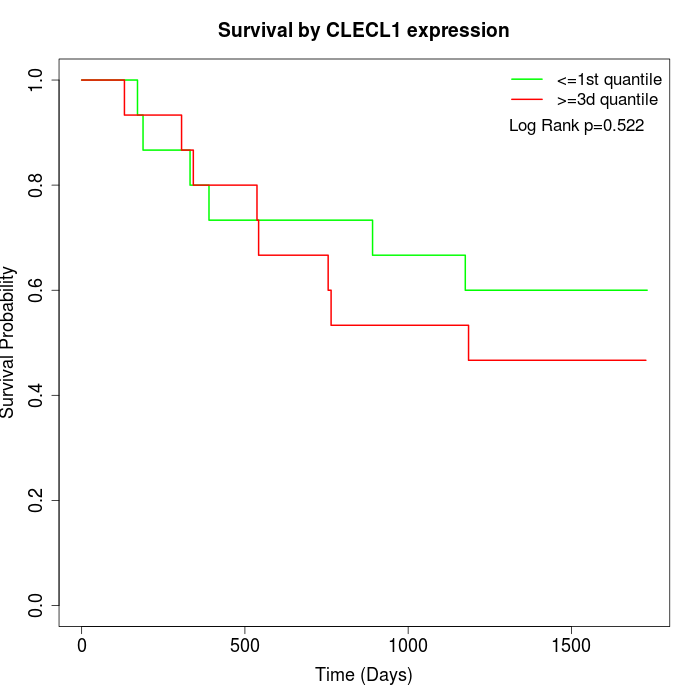
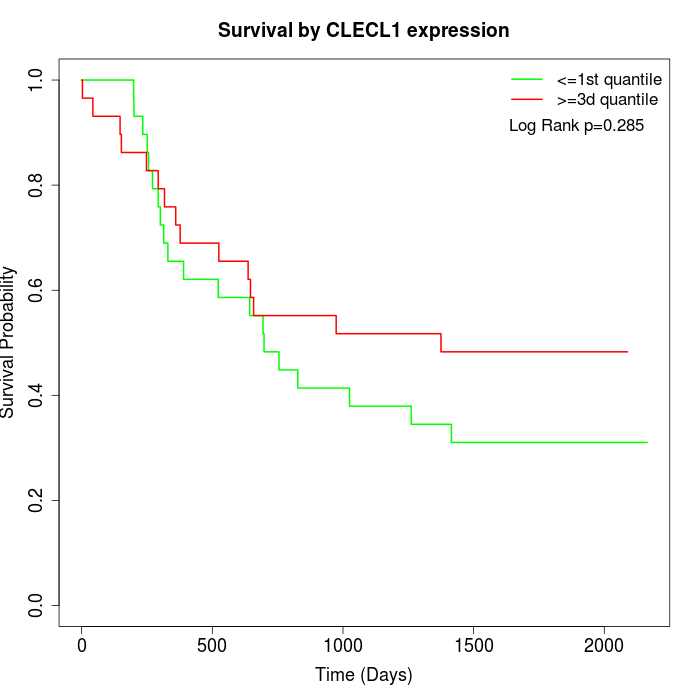
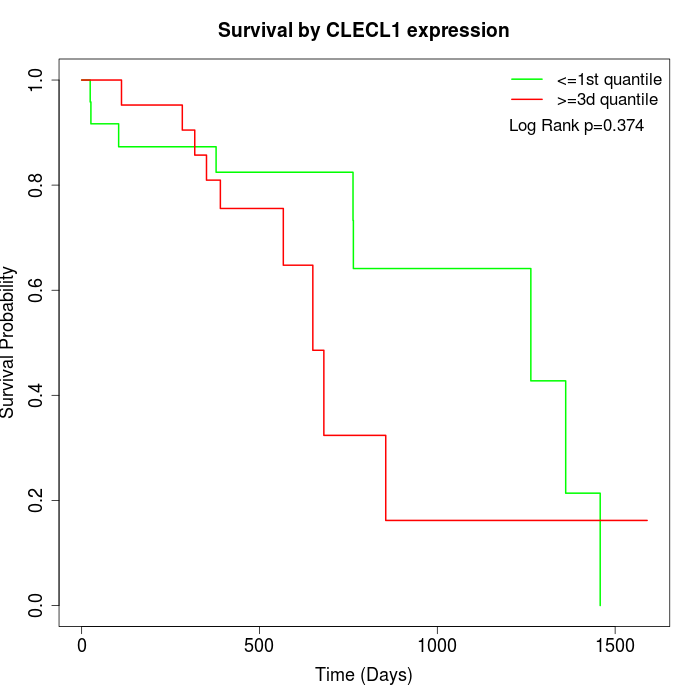
Survival by CLECL1 expression:
Note: Click image to view full size file.
Copy number change of CLECL1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CLECL1 | 160365 | 6 | 3 | 21 | |
| GSE20123 | CLECL1 | 160365 | 6 | 3 | 21 | |
| GSE43470 | CLECL1 | 160365 | 8 | 3 | 32 | |
| GSE46452 | CLECL1 | 160365 | 10 | 1 | 48 | |
| GSE47630 | CLECL1 | 160365 | 14 | 2 | 24 | |
| GSE54993 | CLECL1 | 160365 | 1 | 10 | 59 | |
| GSE54994 | CLECL1 | 160365 | 9 | 2 | 42 | |
| GSE60625 | CLECL1 | 160365 | 0 | 1 | 10 | |
| GSE74703 | CLECL1 | 160365 | 8 | 2 | 26 | |
| GSE74704 | CLECL1 | 160365 | 4 | 2 | 14 | |
| TCGA | CLECL1 | 160365 | 39 | 6 | 51 |
Total number of gains: 105; Total number of losses: 35; Total Number of normals: 348.
Somatic mutations of CLECL1:
Generating mutation plots.
Highly correlated genes for CLECL1:
Showing top 20/165 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CLECL1 | DENND1C | 0.808794 | 3 | 0 | 3 |
| CLECL1 | B3GNTL1 | 0.77531 | 3 | 0 | 3 |
| CLECL1 | PRSS38 | 0.721271 | 3 | 0 | 3 |
| CLECL1 | TRAPPC3L | 0.69705 | 3 | 0 | 3 |
| CLECL1 | OR2B11 | 0.680552 | 3 | 0 | 3 |
| CLECL1 | TRAT1 | 0.677732 | 3 | 0 | 3 |
| CLECL1 | CCDC57 | 0.677028 | 3 | 0 | 3 |
| CLECL1 | MPDU1 | 0.676706 | 3 | 0 | 3 |
| CLECL1 | XAGE2 | 0.676461 | 3 | 0 | 3 |
| CLECL1 | SIGIRR | 0.673006 | 3 | 0 | 3 |
| CLECL1 | DEFB121 | 0.672901 | 4 | 0 | 3 |
| CLECL1 | TXNDC2 | 0.664763 | 4 | 0 | 4 |
| CLECL1 | OR1F1 | 0.662278 | 4 | 0 | 4 |
| CLECL1 | CD27 | 0.661151 | 4 | 0 | 3 |
| CLECL1 | CPNE5 | 0.66 | 5 | 0 | 4 |
| CLECL1 | FAM78A | 0.659052 | 5 | 0 | 3 |
| CLECL1 | IGLL1 | 0.653735 | 6 | 0 | 5 |
| CLECL1 | CD28 | 0.646841 | 4 | 0 | 3 |
| CLECL1 | MAP3K4 | 0.642426 | 3 | 0 | 3 |
| CLECL1 | LINC00996 | 0.640657 | 5 | 0 | 4 |
For details and further investigation, click here