| Full name: ArfGAP with GTPase domain, ankyrin repeat and PH domain 4 | Alias Symbol: Em:AC012044.1|MRIP2 | ||
| Type: protein-coding gene | Cytoband: 10q11.22 | ||
| Entrez ID: 119016 | HGNC ID: HGNC:23459 | Ensembl Gene: ENSG00000188234 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of AGAP4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | AGAP4 | 119016 | 1565620_at | 0.0051 | 0.9966 | |
| GSE26886 | AGAP4 | 119016 | 1565620_at | -0.7523 | 0.0227 | |
| GSE45670 | AGAP4 | 119016 | 1565620_at | 0.3167 | 0.0484 | |
| GSE63941 | AGAP4 | 119016 | 1565620_at | -0.5093 | 0.2120 | |
| GSE77861 | AGAP4 | 119016 | 1565620_at | 0.0706 | 0.4287 | |
| SRP064894 | AGAP4 | 119016 | RNAseq | -0.1298 | 0.6189 | |
| SRP133303 | AGAP4 | 119016 | RNAseq | 0.0117 | 0.9365 | |
| SRP159526 | AGAP4 | 119016 | RNAseq | -0.5458 | 0.0713 | |
| SRP193095 | AGAP4 | 119016 | RNAseq | 0.2181 | 0.1953 | |
| SRP219564 | AGAP4 | 119016 | RNAseq | 0.1483 | 0.6934 | |
| TCGA | AGAP4 | 119016 | RNAseq | -0.2459 | 0.0014 |
Upregulated datasets: 0; Downregulated datasets: 0.
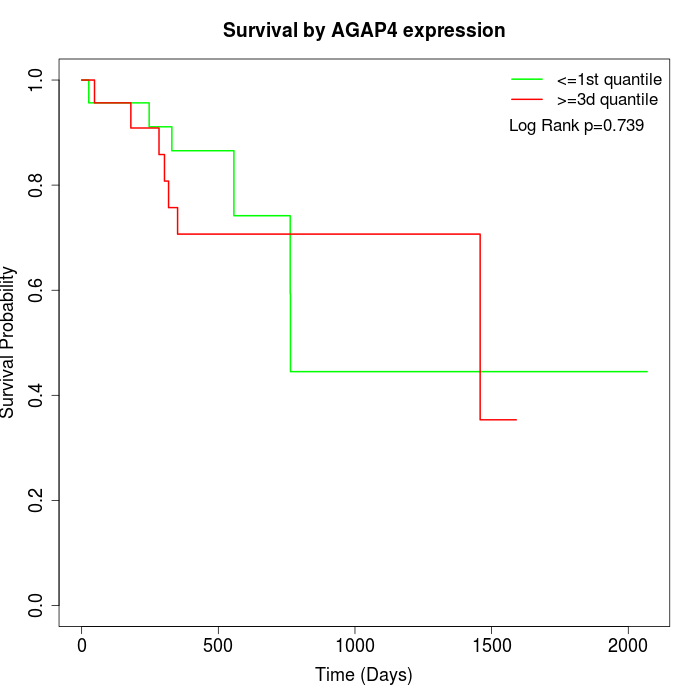
Survival by AGAP4 expression:
Note: Click image to view full size file.
Copy number change of AGAP4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | AGAP4 | 119016 | 3 | 5 | 22 | |
| GSE20123 | AGAP4 | 119016 | 3 | 5 | 22 | |
| GSE43470 | AGAP4 | 119016 | 2 | 6 | 35 | |
| GSE46452 | AGAP4 | 119016 | 0 | 13 | 46 | |
| GSE47630 | AGAP4 | 119016 | 4 | 12 | 24 | |
| GSE54993 | AGAP4 | 119016 | 9 | 0 | 61 | |
| GSE54994 | AGAP4 | 119016 | 2 | 10 | 41 | |
| GSE60625 | AGAP4 | 119016 | 0 | 0 | 11 | |
| GSE74703 | AGAP4 | 119016 | 2 | 3 | 31 | |
| GSE74704 | AGAP4 | 119016 | 2 | 4 | 14 | |
| TCGA | AGAP4 | 119016 | 16 | 18 | 62 |
Total number of gains: 43; Total number of losses: 76; Total Number of normals: 369.
Somatic mutations of AGAP4:
Generating mutation plots.
Highly correlated genes for AGAP4:
Showing top 20/31 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| AGAP4 | GRPEL1 | 0.711522 | 3 | 0 | 3 |
| AGAP4 | HAUS2 | 0.675064 | 3 | 0 | 3 |
| AGAP4 | XRCC2 | 0.660918 | 3 | 0 | 3 |
| AGAP4 | WDPCP | 0.642153 | 3 | 0 | 3 |
| AGAP4 | GTF2H3 | 0.63268 | 4 | 0 | 3 |
| AGAP4 | CCDC152 | 0.632243 | 4 | 0 | 4 |
| AGAP4 | MZT2B | 0.625434 | 4 | 0 | 3 |
| AGAP4 | CDK13 | 0.617614 | 4 | 0 | 3 |
| AGAP4 | ZNF160 | 0.612566 | 4 | 0 | 3 |
| AGAP4 | FBXW12 | 0.609991 | 4 | 0 | 3 |
| AGAP4 | MALAT1 | 0.604774 | 4 | 0 | 4 |
| AGAP4 | CMBL | 0.601824 | 4 | 0 | 3 |
| AGAP4 | NLN | 0.600737 | 4 | 0 | 3 |
| AGAP4 | ZNF721 | 0.596405 | 4 | 0 | 3 |
| AGAP4 | EIF1AD | 0.589417 | 3 | 0 | 3 |
| AGAP4 | DDX59 | 0.582653 | 3 | 0 | 3 |
| AGAP4 | AASDH | 0.581689 | 3 | 0 | 3 |
| AGAP4 | DIP2A | 0.574573 | 5 | 0 | 3 |
| AGAP4 | DBT | 0.564938 | 4 | 0 | 3 |
| AGAP4 | ATF7IP | 0.564433 | 4 | 0 | 3 |
For details and further investigation, click here