| Full name: alanine--glyoxylate aminotransferase 2 | Alias Symbol: AGT2 | ||
| Type: protein-coding gene | Cytoband: 5p13.2 | ||
| Entrez ID: 64902 | HGNC ID: HGNC:14412 | Ensembl Gene: ENSG00000113492 | OMIM ID: 612471 |
| Drug and gene relationship at DGIdb | |||
Expression of AGXT2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | AGXT2 | 64902 | 229229_at | -0.1220 | 0.6529 | |
| GSE26886 | AGXT2 | 64902 | 229229_at | -0.2551 | 0.0467 | |
| GSE45670 | AGXT2 | 64902 | 229229_at | 0.2537 | 0.0096 | |
| GSE53622 | AGXT2 | 64902 | 27208 | 0.0434 | 0.7827 | |
| GSE53624 | AGXT2 | 64902 | 27208 | 0.0624 | 0.3647 | |
| GSE63941 | AGXT2 | 64902 | 229229_at | 0.1160 | 0.4220 | |
| GSE77861 | AGXT2 | 64902 | 229229_at | -0.0095 | 0.9544 | |
| TCGA | AGXT2 | 64902 | RNAseq | 1.3112 | 0.6040 |
Upregulated datasets: 0; Downregulated datasets: 0.
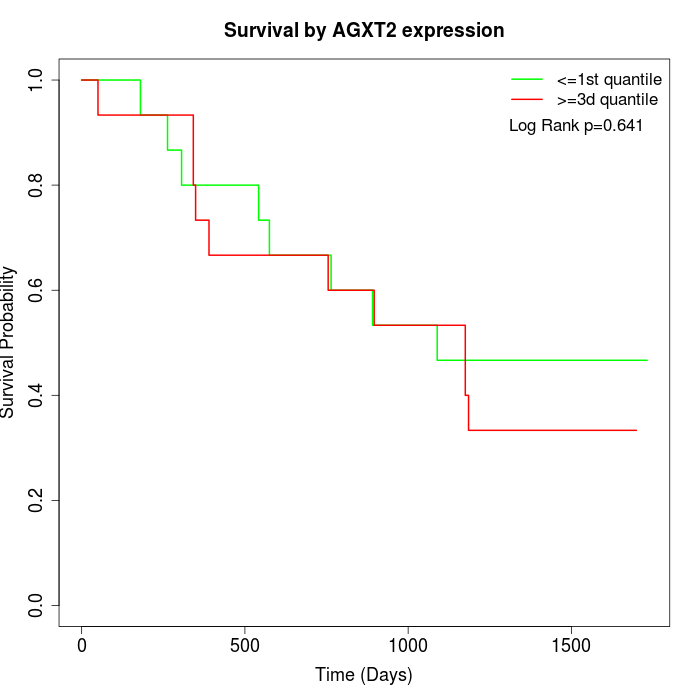
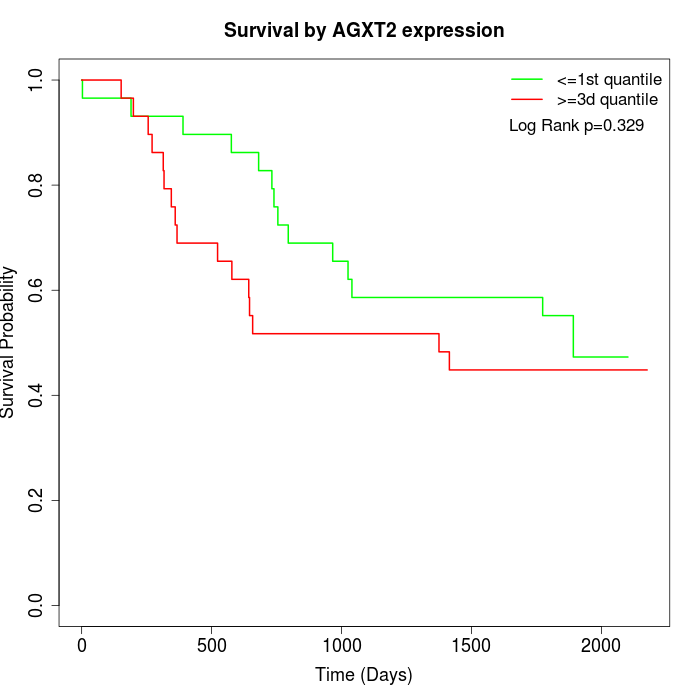
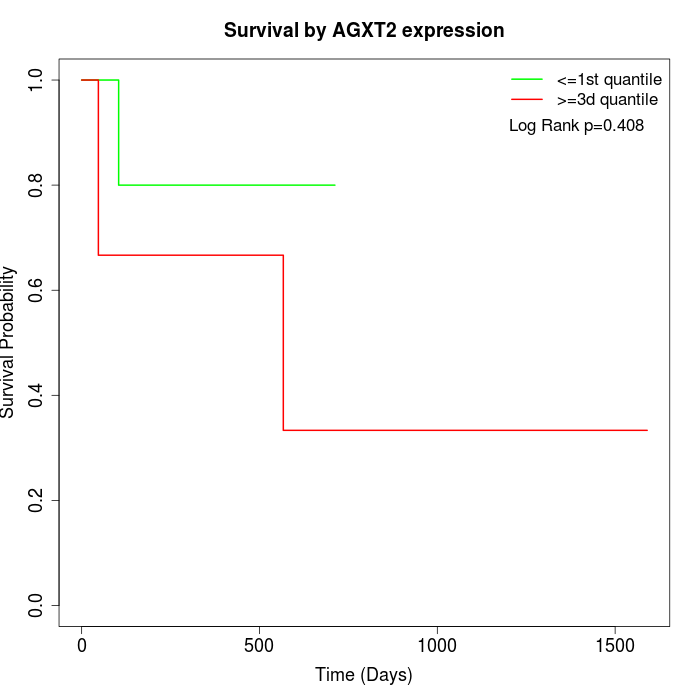
Survival by AGXT2 expression:
Note: Click image to view full size file.
Copy number change of AGXT2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | AGXT2 | 64902 | 10 | 0 | 20 | |
| GSE20123 | AGXT2 | 64902 | 10 | 0 | 20 | |
| GSE43470 | AGXT2 | 64902 | 18 | 0 | 25 | |
| GSE46452 | AGXT2 | 64902 | 6 | 21 | 32 | |
| GSE47630 | AGXT2 | 64902 | 6 | 12 | 22 | |
| GSE54993 | AGXT2 | 64902 | 5 | 4 | 61 | |
| GSE54994 | AGXT2 | 64902 | 26 | 2 | 25 | |
| GSE60625 | AGXT2 | 64902 | 0 | 0 | 11 | |
| GSE74703 | AGXT2 | 64902 | 14 | 0 | 22 | |
| GSE74704 | AGXT2 | 64902 | 9 | 0 | 11 | |
| TCGA | AGXT2 | 64902 | 52 | 6 | 38 |
Total number of gains: 156; Total number of losses: 45; Total Number of normals: 287.
Somatic mutations of AGXT2:
Generating mutation plots.
Highly correlated genes for AGXT2:
Showing top 20/60 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| AGXT2 | CELF2-AS1 | 0.660785 | 3 | 0 | 3 |
| AGXT2 | ASB17 | 0.651524 | 3 | 0 | 3 |
| AGXT2 | CCDC172 | 0.629942 | 4 | 0 | 4 |
| AGXT2 | NCR1 | 0.625328 | 3 | 0 | 3 |
| AGXT2 | PRB4 | 0.624128 | 3 | 0 | 3 |
| AGXT2 | VIPR2 | 0.623798 | 3 | 0 | 3 |
| AGXT2 | ADRB3 | 0.621737 | 4 | 0 | 4 |
| AGXT2 | SETD5 | 0.611851 | 3 | 0 | 3 |
| AGXT2 | ADPRH | 0.609592 | 3 | 0 | 3 |
| AGXT2 | SORCS3-AS1 | 0.609075 | 3 | 0 | 3 |
| AGXT2 | FANCD2OS | 0.608517 | 3 | 0 | 3 |
| AGXT2 | GRIN3A | 0.608483 | 3 | 0 | 3 |
| AGXT2 | ACVR2B-AS1 | 0.607937 | 3 | 0 | 3 |
| AGXT2 | TMEM74 | 0.595637 | 3 | 0 | 3 |
| AGXT2 | SNX32 | 0.590675 | 5 | 0 | 4 |
| AGXT2 | L1CAM | 0.585281 | 4 | 0 | 3 |
| AGXT2 | RANBP3L | 0.581476 | 4 | 0 | 3 |
| AGXT2 | FRMPD3 | 0.581465 | 4 | 0 | 3 |
| AGXT2 | GKN2 | 0.575389 | 4 | 0 | 3 |
| AGXT2 | LINC00951 | 0.574887 | 3 | 0 | 3 |
For details and further investigation, click here