| Full name: autophagy and beclin 1 regulator 1 | Alias Symbol: FLJ20294|KIAA1736|WDR94|DCAF3 | ||
| Type: protein-coding gene | Cytoband: 11p11.2 | ||
| Entrez ID: 55626 | HGNC ID: HGNC:25990 | Ensembl Gene: ENSG00000110497 | OMIM ID: 611359 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of AMBRA1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | AMBRA1 | 55626 | 219141_s_at | -0.0943 | 0.6266 | |
| GSE20347 | AMBRA1 | 55626 | 219141_s_at | -0.1071 | 0.1113 | |
| GSE23400 | AMBRA1 | 55626 | 219141_s_at | -0.1155 | 0.0010 | |
| GSE26886 | AMBRA1 | 55626 | 219141_s_at | -0.2614 | 0.1210 | |
| GSE29001 | AMBRA1 | 55626 | 52731_at | -0.0432 | 0.8978 | |
| GSE38129 | AMBRA1 | 55626 | 219141_s_at | -0.1401 | 0.0394 | |
| GSE45670 | AMBRA1 | 55626 | 219141_s_at | 0.0329 | 0.7525 | |
| GSE53622 | AMBRA1 | 55626 | 121002 | -0.0970 | 0.1264 | |
| GSE53624 | AMBRA1 | 55626 | 58153 | -0.1961 | 0.1001 | |
| GSE63941 | AMBRA1 | 55626 | 219141_s_at | -0.0379 | 0.9022 | |
| GSE77861 | AMBRA1 | 55626 | 219141_s_at | -0.1919 | 0.1096 | |
| GSE97050 | AMBRA1 | 55626 | A_33_P3293169 | -0.2346 | 0.3980 | |
| SRP007169 | AMBRA1 | 55626 | RNAseq | -0.9627 | 0.0516 | |
| SRP008496 | AMBRA1 | 55626 | RNAseq | -0.7609 | 0.0180 | |
| SRP064894 | AMBRA1 | 55626 | RNAseq | -0.2471 | 0.0385 | |
| SRP133303 | AMBRA1 | 55626 | RNAseq | -0.0856 | 0.4859 | |
| SRP159526 | AMBRA1 | 55626 | RNAseq | -0.1792 | 0.5634 | |
| SRP193095 | AMBRA1 | 55626 | RNAseq | -0.3177 | 0.0281 | |
| SRP219564 | AMBRA1 | 55626 | RNAseq | -0.0205 | 0.9165 | |
| TCGA | AMBRA1 | 55626 | RNAseq | -0.0569 | 0.2521 |
Upregulated datasets: 0; Downregulated datasets: 0.
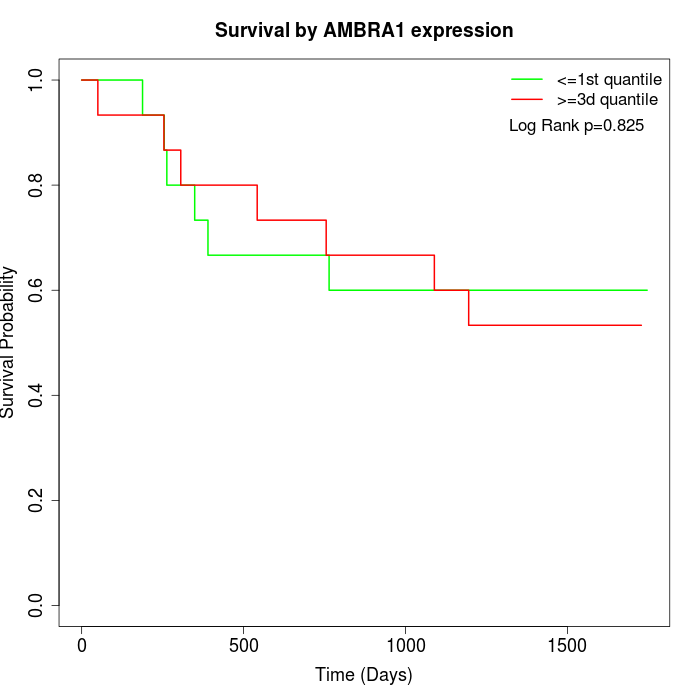
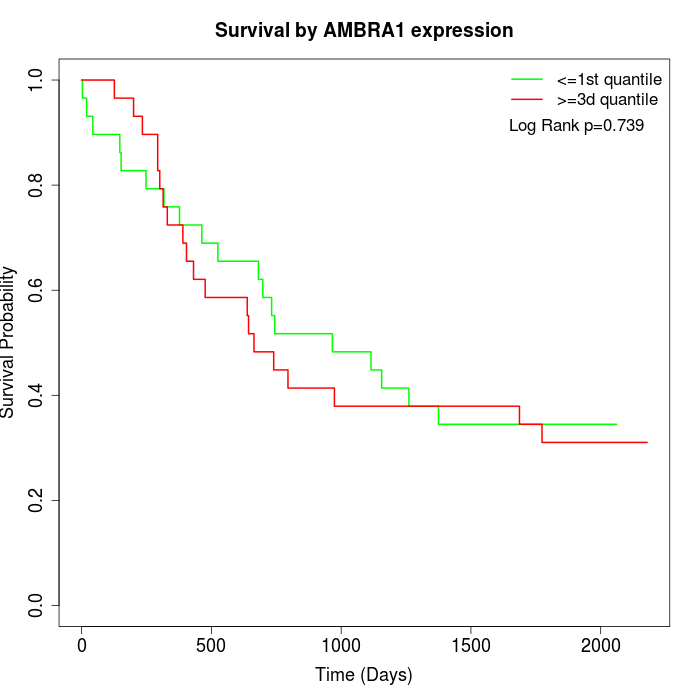
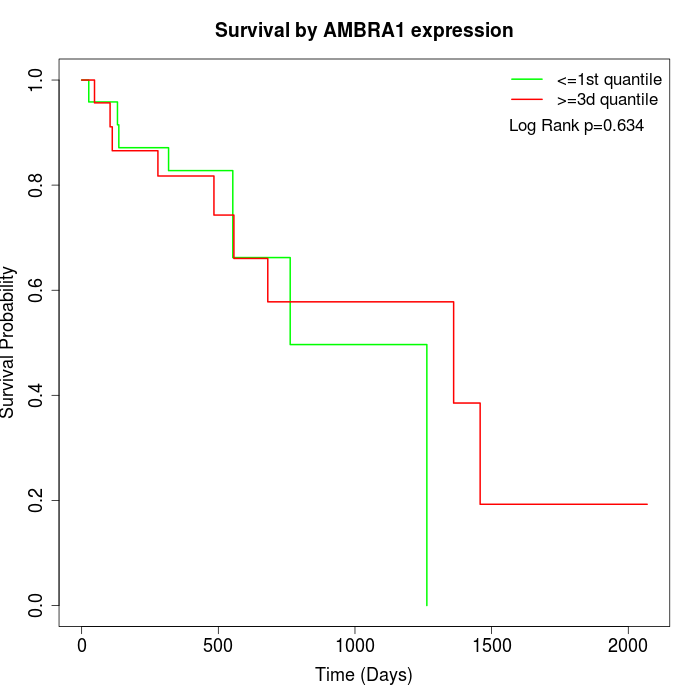
Survival by AMBRA1 expression:
Note: Click image to view full size file.
Copy number change of AMBRA1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | AMBRA1 | 55626 | 3 | 7 | 20 | |
| GSE20123 | AMBRA1 | 55626 | 3 | 6 | 21 | |
| GSE43470 | AMBRA1 | 55626 | 2 | 5 | 36 | |
| GSE46452 | AMBRA1 | 55626 | 8 | 5 | 46 | |
| GSE47630 | AMBRA1 | 55626 | 3 | 9 | 28 | |
| GSE54993 | AMBRA1 | 55626 | 3 | 0 | 67 | |
| GSE54994 | AMBRA1 | 55626 | 1 | 10 | 42 | |
| GSE60625 | AMBRA1 | 55626 | 0 | 0 | 11 | |
| GSE74703 | AMBRA1 | 55626 | 2 | 2 | 32 | |
| GSE74704 | AMBRA1 | 55626 | 2 | 4 | 14 | |
| TCGA | AMBRA1 | 55626 | 14 | 20 | 62 |
Total number of gains: 41; Total number of losses: 68; Total Number of normals: 379.
Somatic mutations of AMBRA1:
Generating mutation plots.
Highly correlated genes for AMBRA1:
Showing top 20/454 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| AMBRA1 | SLC35A4 | 0.771983 | 3 | 0 | 3 |
| AMBRA1 | RNF31 | 0.760396 | 3 | 0 | 3 |
| AMBRA1 | NPFFR1 | 0.757683 | 4 | 0 | 3 |
| AMBRA1 | ZBTB33 | 0.744849 | 3 | 0 | 3 |
| AMBRA1 | TP53BP2 | 0.729687 | 3 | 0 | 3 |
| AMBRA1 | TSPO | 0.723417 | 4 | 0 | 4 |
| AMBRA1 | CRB3 | 0.721991 | 3 | 0 | 3 |
| AMBRA1 | NCOA1 | 0.717866 | 3 | 0 | 3 |
| AMBRA1 | NPBWR2 | 0.715972 | 3 | 0 | 3 |
| AMBRA1 | MAP1A | 0.714235 | 3 | 0 | 3 |
| AMBRA1 | NANOG | 0.713565 | 3 | 0 | 3 |
| AMBRA1 | KALRN | 0.712766 | 4 | 0 | 4 |
| AMBRA1 | ATF7 | 0.707142 | 3 | 0 | 3 |
| AMBRA1 | PIM1 | 0.706897 | 3 | 0 | 3 |
| AMBRA1 | EIF1AY | 0.706172 | 3 | 0 | 3 |
| AMBRA1 | KEL | 0.705674 | 3 | 0 | 3 |
| AMBRA1 | UFM1 | 0.704459 | 3 | 0 | 3 |
| AMBRA1 | KLHDC10 | 0.703676 | 3 | 0 | 3 |
| AMBRA1 | TLR5 | 0.703605 | 4 | 0 | 4 |
| AMBRA1 | CYP2B6 | 0.703516 | 3 | 0 | 3 |
For details and further investigation, click here