| Full name: nuclear receptor coactivator 1 | Alias Symbol: SRC1|F-SRC-1|NCoA-1|KAT13A|RIP160|bHLHe74 | ||
| Type: protein-coding gene | Cytoband: 2p23.3 | ||
| Entrez ID: 8648 | HGNC ID: HGNC:7668 | Ensembl Gene: ENSG00000084676 | OMIM ID: 602691 |
| Drug and gene relationship at DGIdb | |||
Expression of NCOA1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | NCOA1 | 8648 | 209107_x_at | -0.6436 | 0.0502 | |
| GSE20347 | NCOA1 | 8648 | 210249_s_at | -0.8330 | 0.0000 | |
| GSE23400 | NCOA1 | 8648 | 209107_x_at | -0.5970 | 0.0000 | |
| GSE26886 | NCOA1 | 8648 | 210249_s_at | 0.1667 | 0.5361 | |
| GSE29001 | NCOA1 | 8648 | 209106_at | -1.3200 | 0.0012 | |
| GSE38129 | NCOA1 | 8648 | 209107_x_at | -0.7993 | 0.0000 | |
| GSE45670 | NCOA1 | 8648 | 209107_x_at | -0.4579 | 0.0244 | |
| GSE53622 | NCOA1 | 8648 | 123744 | -0.9206 | 0.0000 | |
| GSE53624 | NCOA1 | 8648 | 123744 | -1.2267 | 0.0000 | |
| GSE63941 | NCOA1 | 8648 | 209107_x_at | -0.0869 | 0.8821 | |
| GSE77861 | NCOA1 | 8648 | 209107_x_at | -0.5181 | 0.0332 | |
| GSE97050 | NCOA1 | 8648 | A_23_P39602 | -0.4352 | 0.1889 | |
| SRP007169 | NCOA1 | 8648 | RNAseq | -1.2794 | 0.0089 | |
| SRP008496 | NCOA1 | 8648 | RNAseq | -1.0091 | 0.0005 | |
| SRP064894 | NCOA1 | 8648 | RNAseq | -1.1889 | 0.0000 | |
| SRP133303 | NCOA1 | 8648 | RNAseq | -0.7069 | 0.0000 | |
| SRP159526 | NCOA1 | 8648 | RNAseq | -0.4213 | 0.1437 | |
| SRP193095 | NCOA1 | 8648 | RNAseq | -0.7287 | 0.0000 | |
| SRP219564 | NCOA1 | 8648 | RNAseq | -0.5593 | 0.1333 | |
| TCGA | NCOA1 | 8648 | RNAseq | -0.2096 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 5.
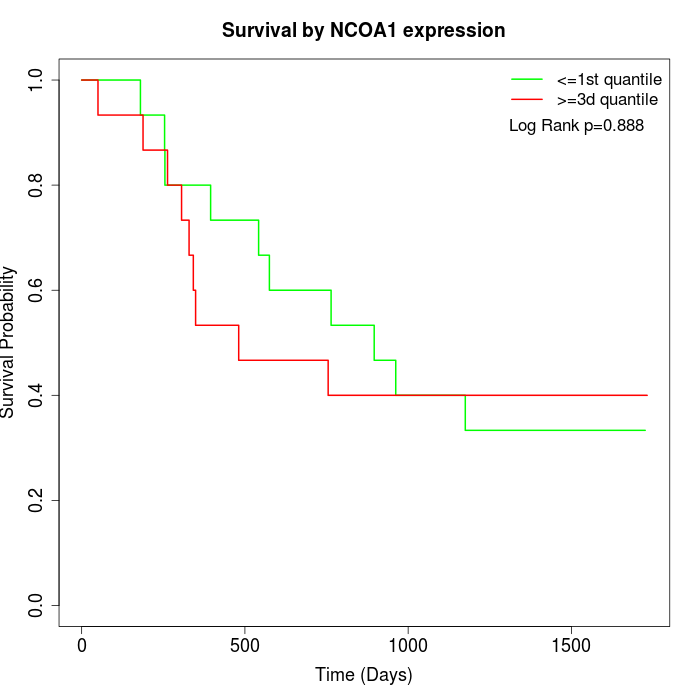
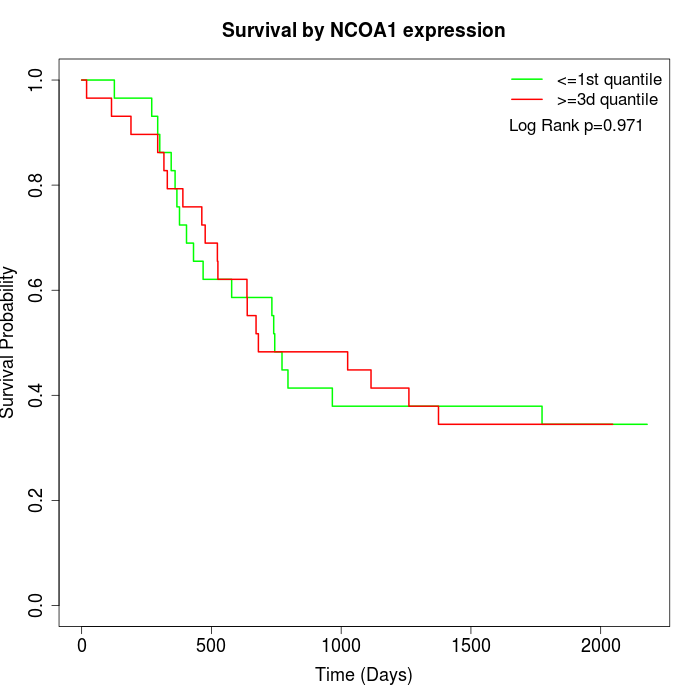
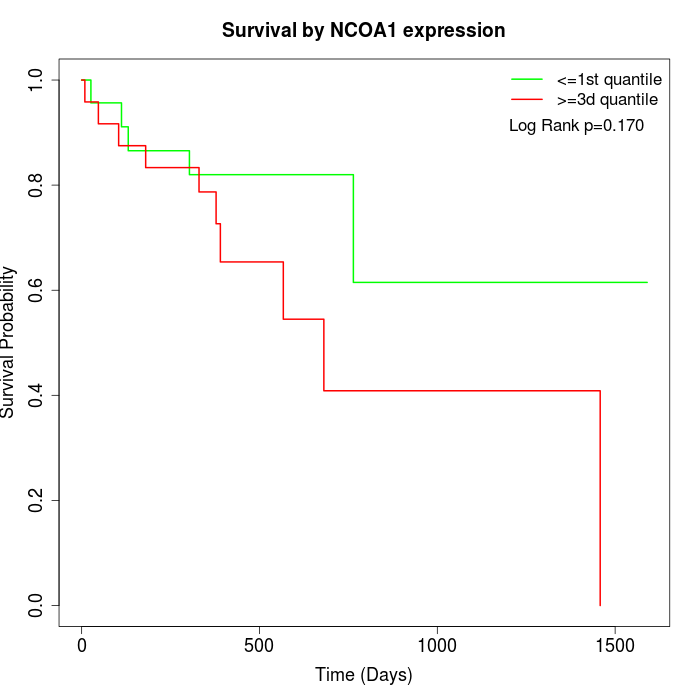
Survival by NCOA1 expression:
Note: Click image to view full size file.
Copy number change of NCOA1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | NCOA1 | 8648 | 11 | 2 | 17 | |
| GSE20123 | NCOA1 | 8648 | 11 | 2 | 17 | |
| GSE43470 | NCOA1 | 8648 | 2 | 1 | 40 | |
| GSE46452 | NCOA1 | 8648 | 3 | 4 | 52 | |
| GSE47630 | NCOA1 | 8648 | 8 | 0 | 32 | |
| GSE54993 | NCOA1 | 8648 | 0 | 6 | 64 | |
| GSE54994 | NCOA1 | 8648 | 10 | 0 | 43 | |
| GSE60625 | NCOA1 | 8648 | 0 | 3 | 8 | |
| GSE74703 | NCOA1 | 8648 | 2 | 0 | 34 | |
| GSE74704 | NCOA1 | 8648 | 9 | 1 | 10 | |
| TCGA | NCOA1 | 8648 | 36 | 4 | 56 |
Total number of gains: 92; Total number of losses: 23; Total Number of normals: 373.
Somatic mutations of NCOA1:
Generating mutation plots.
Highly correlated genes for NCOA1:
Showing top 20/1539 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| NCOA1 | RMND5B | 0.822657 | 10 | 0 | 10 |
| NCOA1 | SNORA68 | 0.78061 | 3 | 0 | 3 |
| NCOA1 | KIAA1211L | 0.779836 | 5 | 0 | 5 |
| NCOA1 | CGNL1 | 0.774084 | 6 | 0 | 6 |
| NCOA1 | ARHGEF10L | 0.772637 | 10 | 0 | 10 |
| NCOA1 | FCHO2 | 0.772296 | 6 | 0 | 6 |
| NCOA1 | KIAA0232 | 0.772205 | 9 | 0 | 9 |
| NCOA1 | ACOX3 | 0.771492 | 9 | 0 | 9 |
| NCOA1 | YPEL3 | 0.769101 | 7 | 0 | 6 |
| NCOA1 | FLG-AS1 | 0.768424 | 4 | 0 | 4 |
| NCOA1 | SFTA2 | 0.767209 | 3 | 0 | 3 |
| NCOA1 | SMIM5 | 0.767093 | 5 | 0 | 5 |
| NCOA1 | GALNT12 | 0.764837 | 10 | 0 | 10 |
| NCOA1 | ZNF503-AS1 | 0.763366 | 5 | 0 | 5 |
| NCOA1 | TRNP1 | 0.76156 | 5 | 0 | 5 |
| NCOA1 | BEX4 | 0.76056 | 10 | 0 | 10 |
| NCOA1 | KRT78 | 0.760313 | 5 | 0 | 5 |
| NCOA1 | PLEKHA7 | 0.759687 | 6 | 0 | 6 |
| NCOA1 | SASH1 | 0.758757 | 10 | 0 | 10 |
| NCOA1 | SORT1 | 0.754921 | 10 | 0 | 10 |
For details and further investigation, click here