| Full name: amyloid beta precursor protein | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 21q21.3 | ||
| Entrez ID: 351 | HGNC ID: HGNC:620 | Ensembl Gene: ENSG00000142192 | OMIM ID: 104760 |
| Drug and gene relationship at DGIdb | |||
APP involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05010 | Alzheimer's disease |
Expression of APP:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | APP | 351 | 214953_s_at | 0.5984 | 0.2654 | |
| GSE20347 | APP | 351 | 200602_at | 0.9191 | 0.0000 | |
| GSE23400 | APP | 351 | 214953_s_at | 0.7221 | 0.0000 | |
| GSE26886 | APP | 351 | 200602_at | 1.2165 | 0.0003 | |
| GSE29001 | APP | 351 | 200602_at | 0.8462 | 0.0005 | |
| GSE38129 | APP | 351 | 200602_at | 0.4849 | 0.0193 | |
| GSE45670 | APP | 351 | 214953_s_at | 0.3075 | 0.2323 | |
| GSE53622 | APP | 351 | 54920 | 0.5317 | 0.0000 | |
| GSE53624 | APP | 351 | 54920 | 0.7882 | 0.0000 | |
| GSE63941 | APP | 351 | 214953_s_at | -0.6121 | 0.3179 | |
| GSE77861 | APP | 351 | 200602_at | 0.5411 | 0.2735 | |
| GSE97050 | APP | 351 | A_33_P3296479 | 0.0079 | 0.9808 | |
| SRP007169 | APP | 351 | RNAseq | 0.4131 | 0.2727 | |
| SRP008496 | APP | 351 | RNAseq | 1.1794 | 0.0036 | |
| SRP064894 | APP | 351 | RNAseq | 0.9931 | 0.0000 | |
| SRP133303 | APP | 351 | RNAseq | 0.5909 | 0.0010 | |
| SRP159526 | APP | 351 | RNAseq | 1.1205 | 0.0001 | |
| SRP193095 | APP | 351 | RNAseq | 1.2211 | 0.0000 | |
| SRP219564 | APP | 351 | RNAseq | 0.2677 | 0.6489 | |
| TCGA | APP | 351 | RNAseq | 0.0513 | 0.2602 |
Upregulated datasets: 4; Downregulated datasets: 0.
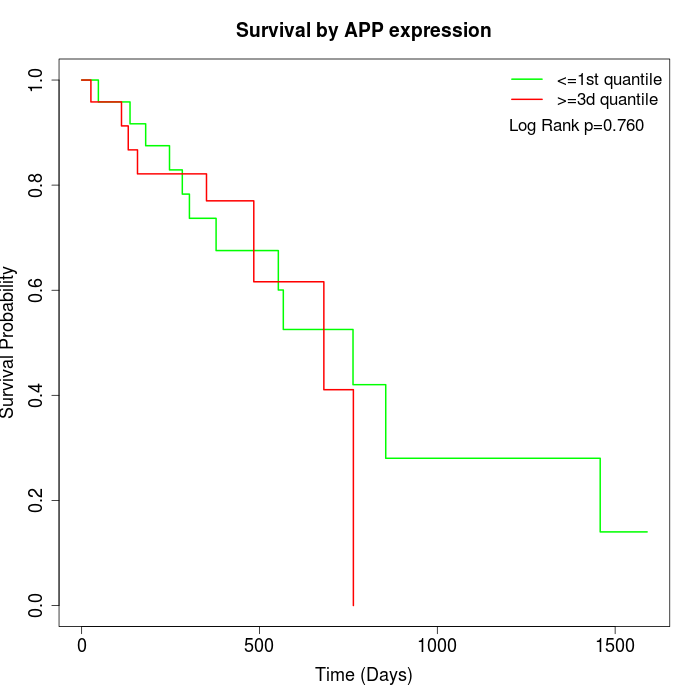
Survival by APP expression:
Note: Click image to view full size file.
Copy number change of APP:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | APP | 351 | 2 | 11 | 17 | |
| GSE20123 | APP | 351 | 2 | 11 | 17 | |
| GSE43470 | APP | 351 | 1 | 8 | 34 | |
| GSE46452 | APP | 351 | 1 | 21 | 37 | |
| GSE47630 | APP | 351 | 6 | 17 | 17 | |
| GSE54993 | APP | 351 | 9 | 2 | 59 | |
| GSE54994 | APP | 351 | 2 | 8 | 43 | |
| GSE60625 | APP | 351 | 0 | 0 | 11 | |
| GSE74703 | APP | 351 | 1 | 6 | 29 | |
| GSE74704 | APP | 351 | 2 | 7 | 11 | |
| TCGA | APP | 351 | 9 | 38 | 49 |
Total number of gains: 35; Total number of losses: 129; Total Number of normals: 324.
Somatic mutations of APP:
Generating mutation plots.
Highly correlated genes for APP:
Showing top 20/1295 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| APP | ASB7 | 0.773591 | 3 | 0 | 3 |
| APP | PET117 | 0.769193 | 4 | 0 | 4 |
| APP | ENAH | 0.739707 | 11 | 0 | 11 |
| APP | DPP9 | 0.7245 | 3 | 0 | 3 |
| APP | BCL9L | 0.723959 | 3 | 0 | 3 |
| APP | PHRF1 | 0.721171 | 3 | 0 | 3 |
| APP | ZNF391 | 0.72109 | 3 | 0 | 3 |
| APP | CHST15 | 0.718741 | 11 | 0 | 10 |
| APP | SLC25A29 | 0.709832 | 3 | 0 | 3 |
| APP | FBLIM1 | 0.708384 | 7 | 0 | 7 |
| APP | SCAF4 | 0.701617 | 3 | 0 | 3 |
| APP | CDKN1C | 0.699741 | 4 | 0 | 3 |
| APP | TIMP4 | 0.69794 | 3 | 0 | 3 |
| APP | TTC7B | 0.696715 | 7 | 0 | 7 |
| APP | B3GNT9 | 0.695078 | 7 | 0 | 7 |
| APP | EIF5B | 0.693107 | 4 | 0 | 4 |
| APP | LTBP1 | 0.693051 | 11 | 0 | 11 |
| APP | SEC22A | 0.691508 | 4 | 0 | 4 |
| APP | LINC00674 | 0.690304 | 3 | 0 | 3 |
| APP | SLC25A30 | 0.687251 | 5 | 0 | 4 |
For details and further investigation, click here