| Full name: ENAH actin regulator | Alias Symbol: FLJ10773|NDPP1|MENA | ||
| Type: protein-coding gene | Cytoband: 1q42.12 | ||
| Entrez ID: 55740 | HGNC ID: HGNC:18271 | Ensembl Gene: ENSG00000154380 | OMIM ID: 609061 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
ENAH involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04360 | Axon guidance | |
| hsa04810 | Regulation of actin cytoskeleton |
Expression of ENAH:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ENAH | 55740 | 222433_at | 1.2462 | 0.0374 | |
| GSE20347 | ENAH | 55740 | 217820_s_at | 1.9621 | 0.0000 | |
| GSE23400 | ENAH | 55740 | 217820_s_at | 1.4253 | 0.0000 | |
| GSE26886 | ENAH | 55740 | 222433_at | 2.5936 | 0.0000 | |
| GSE29001 | ENAH | 55740 | 217820_s_at | 1.9628 | 0.0000 | |
| GSE38129 | ENAH | 55740 | 217820_s_at | 1.4308 | 0.0002 | |
| GSE45670 | ENAH | 55740 | 217820_s_at | 0.9156 | 0.0065 | |
| GSE53622 | ENAH | 55740 | 82856 | 1.0777 | 0.0000 | |
| GSE53624 | ENAH | 55740 | 82856 | 1.4798 | 0.0000 | |
| GSE63941 | ENAH | 55740 | 222433_at | -1.2923 | 0.0137 | |
| GSE77861 | ENAH | 55740 | 222433_at | 2.2060 | 0.0010 | |
| GSE97050 | ENAH | 55740 | A_23_P51397 | 0.1963 | 0.7189 | |
| SRP007169 | ENAH | 55740 | RNAseq | 2.5044 | 0.0000 | |
| SRP008496 | ENAH | 55740 | RNAseq | 2.6476 | 0.0000 | |
| SRP064894 | ENAH | 55740 | RNAseq | 1.0456 | 0.0000 | |
| SRP133303 | ENAH | 55740 | RNAseq | 1.4254 | 0.0000 | |
| SRP159526 | ENAH | 55740 | RNAseq | 1.2251 | 0.0001 | |
| SRP193095 | ENAH | 55740 | RNAseq | 1.7220 | 0.0000 | |
| SRP219564 | ENAH | 55740 | RNAseq | 0.2889 | 0.5619 | |
| TCGA | ENAH | 55740 | RNAseq | 0.0670 | 0.2655 |
Upregulated datasets: 15; Downregulated datasets: 1.
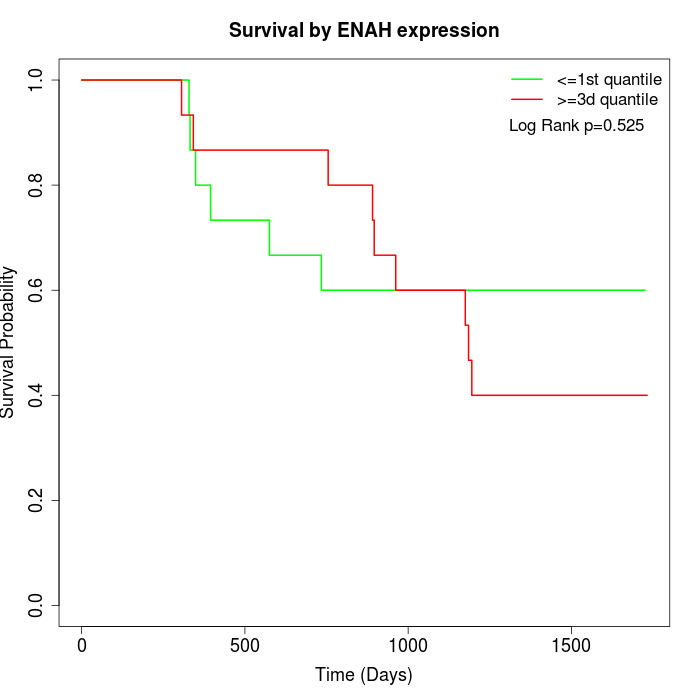
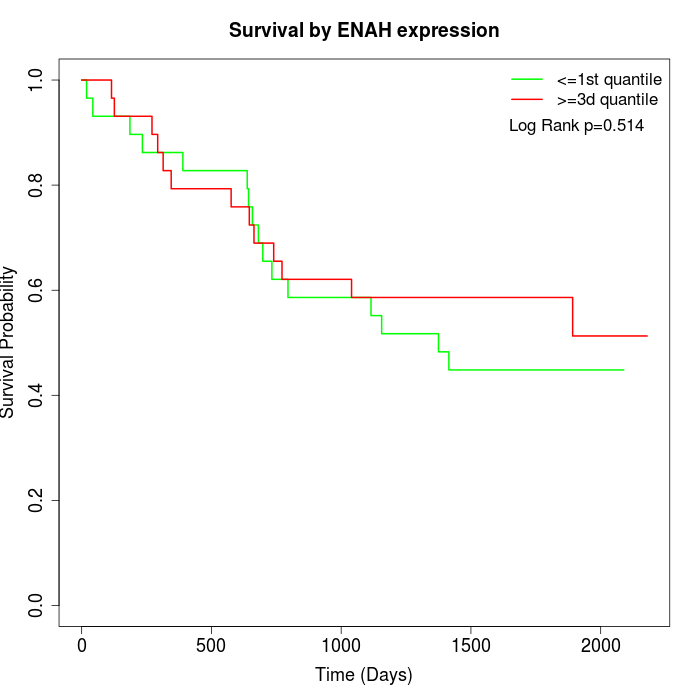
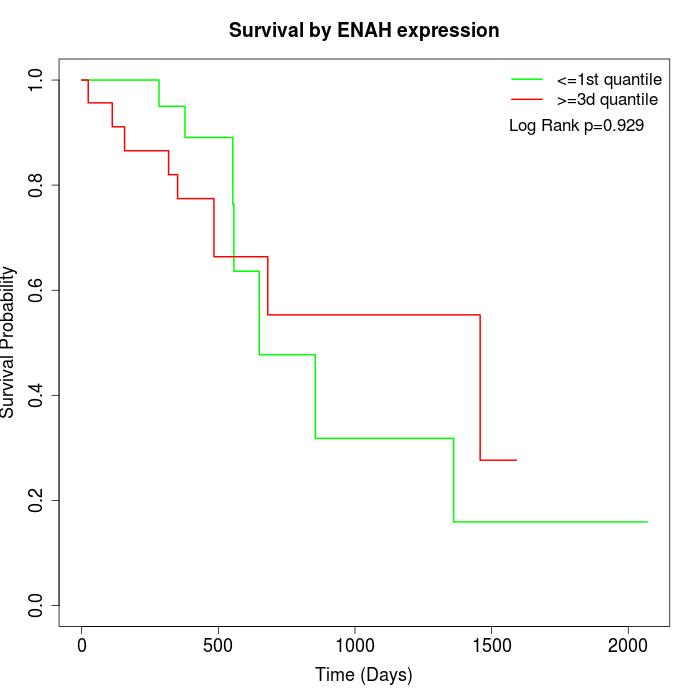
Survival by ENAH expression:
Note: Click image to view full size file.
Copy number change of ENAH:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ENAH | 55740 | 11 | 0 | 19 | |
| GSE20123 | ENAH | 55740 | 11 | 0 | 19 | |
| GSE43470 | ENAH | 55740 | 7 | 1 | 35 | |
| GSE46452 | ENAH | 55740 | 3 | 1 | 55 | |
| GSE47630 | ENAH | 55740 | 15 | 0 | 25 | |
| GSE54993 | ENAH | 55740 | 0 | 6 | 64 | |
| GSE54994 | ENAH | 55740 | 16 | 0 | 37 | |
| GSE60625 | ENAH | 55740 | 0 | 0 | 11 | |
| GSE74703 | ENAH | 55740 | 7 | 1 | 28 | |
| GSE74704 | ENAH | 55740 | 5 | 0 | 15 | |
| TCGA | ENAH | 55740 | 46 | 3 | 47 |
Total number of gains: 121; Total number of losses: 12; Total Number of normals: 355.
Somatic mutations of ENAH:
Generating mutation plots.
Highly correlated genes for ENAH:
Showing top 20/1795 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ENAH | SERPINH1 | 0.808683 | 10 | 0 | 10 |
| ENAH | SEMA3C | 0.798295 | 11 | 0 | 11 |
| ENAH | INHBA | 0.797028 | 11 | 0 | 10 |
| ENAH | FADS1 | 0.796471 | 3 | 0 | 3 |
| ENAH | COL4A1 | 0.785498 | 12 | 0 | 12 |
| ENAH | LGALS1 | 0.784619 | 11 | 0 | 11 |
| ENAH | FNDC3B | 0.777556 | 11 | 0 | 11 |
| ENAH | LPCAT1 | 0.770578 | 12 | 0 | 11 |
| ENAH | TGFBI | 0.766593 | 12 | 0 | 11 |
| ENAH | ITGAV | 0.76612 | 12 | 0 | 11 |
| ENAH | SLC39A14 | 0.765192 | 12 | 0 | 11 |
| ENAH | FMNL2 | 0.763332 | 8 | 0 | 8 |
| ENAH | MSN | 0.762688 | 12 | 0 | 11 |
| ENAH | FBLIM1 | 0.762023 | 8 | 0 | 7 |
| ENAH | ASAP1 | 0.757685 | 12 | 0 | 12 |
| ENAH | PRNP | 0.751513 | 12 | 0 | 11 |
| ENAH | COL4A2 | 0.747735 | 12 | 0 | 12 |
| ENAH | PLAU | 0.746604 | 11 | 0 | 11 |
| ENAH | MMD | 0.74538 | 12 | 0 | 12 |
| ENAH | NFE2L1 | 0.74464 | 11 | 0 | 11 |
For details and further investigation, click here